3BWZ
 
 | | Crystal structure of the type II cohesin module from the cellulosome of Acetivibrio cellulolyticus with an extended linker conformation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Lamed, R, Shimon, L.J.W, Bayer, E, Frolow, F. | | Deposit date: | 2008-01-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
4UYQ
 
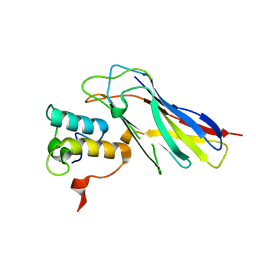 | |
8X3A
 
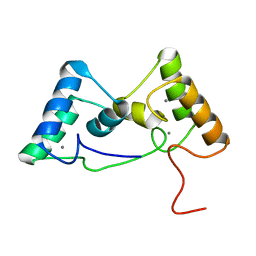 | |
8X39
 
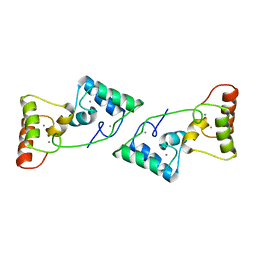 | |
4N2O
 
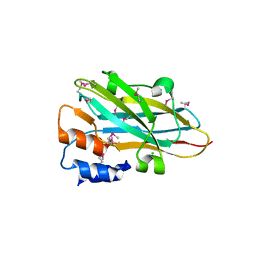 | | Structure of a novel autonomous cohesin protein from Ruminococcus flavefaciens | | Descriptor: | Autonomous cohesin, CHLORIDE ION | | Authors: | Frolow, F, Voronov-Goldman, M, Levy-Assaraf, M, Lamed, R, Bayer, E, Shimon, L. | | Deposit date: | 2013-10-05 | | Release date: | 2013-12-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural characterization of a novel autonomous cohesin from Ruminococcus flavefaciens.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8HDJ
 
 | |
8HER
 
 | |
8HEP
 
 | |
8HEQ
 
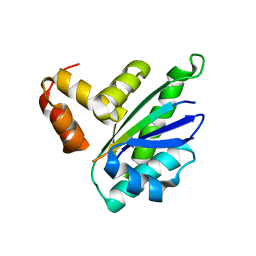 | |
7RPY
 
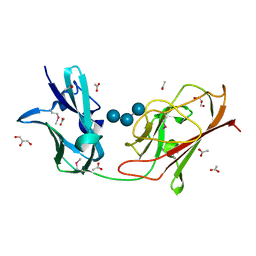 | | X25-2 domain of Sca5 from Ruminococcus bromii | | Descriptor: | ACETATE ION, Cohesin-containing protein, GLYCEROL, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome.
J.Biol.Chem., 298, 2022
|
|
7RFT
 
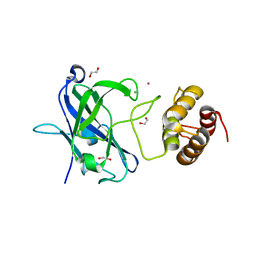 | |
7RAW
 
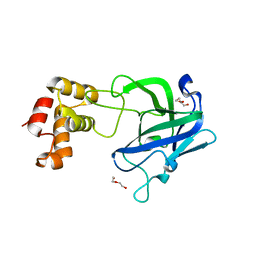 | |
5LXV
 
 | |
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
4UMS
 
 | | The crystal structure of the seventh ScaB type I cohesin from Pseudobacteroides cellulosolvens | | Descriptor: | CELLULOSOMAL ANCHORING SCAFFOLDIN B | | Authors: | Cameron, K, Alves, V.D, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Combined Crystal Structure of a Type-I Cohesin, Mutation and Affinity-Binding Studies Reveal Structural Determinants of Cohesin-Dockerin Specificity
J.Biol.Chem., 290, 2015
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
1NBC
 
 | | BACTERIAL TYPE 3A CELLULOSE-BINDING DOMAIN | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A | | Authors: | Tormo, J, Lamed, R, Steitz, T.A. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose.
EMBO J., 15, 1996
|
|
4Z28
 
 | | Crystal structure of short hoefavidin biotin complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avidin family, BIOTIN | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hoefavidin: A dimeric bacterial avidin with a C-terminal binding tail.
J.Struct.Biol., 191, 2015
|
|
4Z6J
 
 | |
3ZUC
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus determined from the crystals grown in the presence of Nickel | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-18 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ZU8
 
 | | STRUCTURE OF CBM3B OF MAJOR SCAFFOLDIN SUBUNIT SCAA FROM ACETIVIBRIO CELLULOLYTICUS DETERMINED ON THE NIKEL ABSORPTION EDGE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ZQW
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4Z2V
 
 | |
