1AIP
 
 | | EF-TU EF-TS COMPLEX FROM THERMUS THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS, ELONGATION FACTOR TU | | Authors: | Wang, Y, Jiang, Y, Meyering-Voss, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1997-04-22 | | Release date: | 1997-10-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
1A52
 
 | |
1AIS
 
 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*AP*CP*TP*TP*TP*(5IU)P*(5IU)P*AP*AP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*AP*AP*GP*TP*T )-3'), PROTEIN (TATA-BINDING PROTEIN), ... | | Authors: | Kosa, P.F, Ghosh, G, Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1997-04-24 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1BUN
 
 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1QVR
 
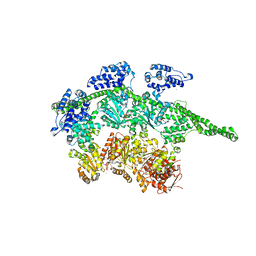 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
1POD
 
 | |
1R4R
 
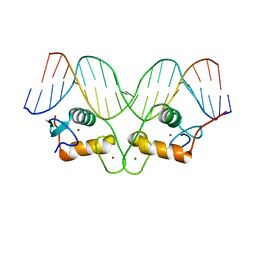 | | Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*GP*AP*AP*CP*AP*TP*CP*AP*TP*GP*TP*TP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*TP*GP*AP*TP*GP*TP*TP*CP*TP*CP*A)-3', Glucocorticoid receptor, ... | | Authors: | Luisi, B.F, Xu, W.X, Otwinowski, Z, Freedman, L.P, Yamamoto, K.R, Sigler, P.B. | | Deposit date: | 2003-10-07 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic Analysis of the Interaction of the Glucocorticoid Receptor with DNA
Nature, 352, 1991
|
|
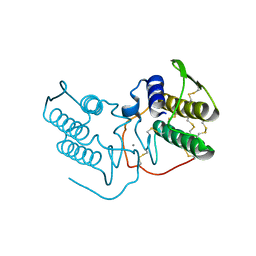
1TFE
 
 | | DIMERIZATION DOMAIN OF EF-TS FROM T. THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS | | Authors: | Jiang, Y, Nock, S, Nesper, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus.
Biochemistry, 35, 1996
|
|
1PCZ
 
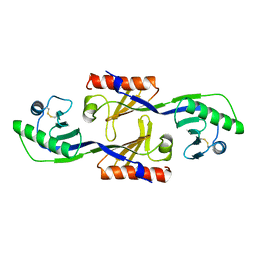 | | STRUCTURE OF TATA-BINDING PROTEIN | | Descriptor: | TATA-BINDING PROTEIN | | Authors: | Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1996-10-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a hyperthermophilic archaeal TATA-box binding protein.
J.Mol.Biol., 264, 1996
|
|
1PF9
 
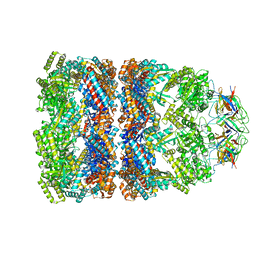 | | GroEL-GroES-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, groEL protein, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-24 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
1PCQ
 
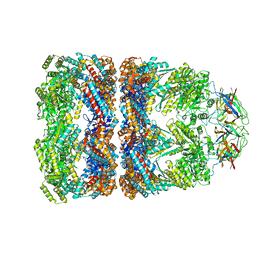 | | Crystal structure of groEL-groES | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-16 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
4V43
 
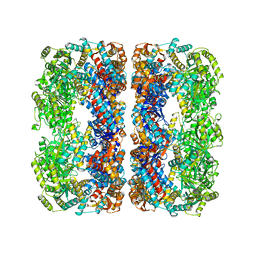 | |
1MNF
 
 | |
4CHA
 
 | |
1GR5
 
 | | Solution Structure of apo GroEL by Cryo-Electron microscopy | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1KP8
 
 | |
2CHA
 
 | |
4V8S
 
 | | Archaeal RNAP-DNA binary complex at 4.32Ang | | Descriptor: | 5'-D(*AP*TP*AP*GP*AP*GP*TP*AP*TP*AP*AP*GP*AP*TP *AP*G)-3', 5'-D(*TP*CP*TP*TP*AP*TP*AP*CP*TP*CP*TP*AP*TP*CP)-3', DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-07-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.323 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
1VAP
 
 | |
2GDA
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1G4R
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN 1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
1G4M
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
4AYB
 
 | | RNAP at 3.2Ang | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
2Y0S
 
 | | Crystal structure of Sulfolobus shibatae RNA polymerase in P21 space group | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, DNA-DIRECTED RNA POLYMERASE SUBUNIT A'', DNA-DIRECTED RNA POLYMERASE SUBUNIT D, ... | | Authors: | Wojtas, M, Peralta, B, Ondiviela, M, Mogni, M, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Archaeal RNA polymerase: the influence of the protruding stalk in crystal packing and preliminary biophysical analysis of the Rpo13 subunit.
Biochem. Soc. Trans., 39, 2011
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
