7VS7
 
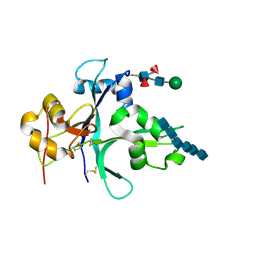 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
7DJJ
 
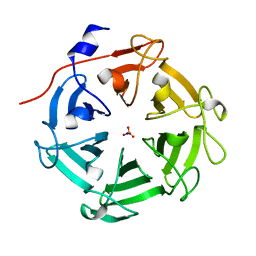 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
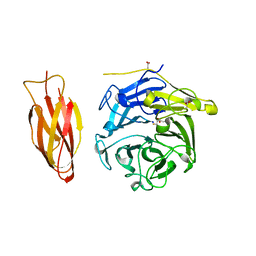 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
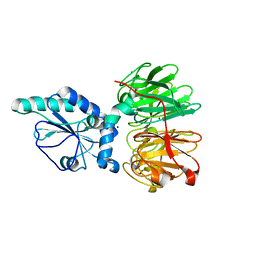 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7E5C
 
 | | Bacterial prolidase mutant D45W/L225Y/H226L/H343I | | Descriptor: | MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Yang, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis
Catalysis Science And Technology, 11, 2021
|
|
7Y1A
 
 | | Lateral hexamer | | Descriptor: | B-phycoerythrin beta chain, LRH, PHYCOERYTHROBILIN, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y4L
 
 | | PBS of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta 18 subunit, Allophycocyanin beta subunit, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-15 | | Release date: | 2023-01-18 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7CRZ
 
 | | Crystal structure of human glucose transporter GLUT3 bound with C3361 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Solute carrier family 2, ... | | Authors: | Yuan, Y.Y, Zhang, S, Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orthosteric-allosteric dual inhibitors of PfHT1 as selective antimalarial agents.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XPR
 
 | | Crystal structrue of SeMet-MtdL:GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPV
 
 | | crysteal structure of MtdL-S228A-His soaked with GDP-Fucf and Mn | | Descriptor: | MANGANESE (II) ION, Transglycosylse, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{S},3~{S},4~{S},5~{R})-3,4-bis(oxidanyl)-5-[(1~{S})-1-oxidanylethyl]oxolan-2-yl] hydrogen phosphate | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPT
 
 | | Crystal structrue of MtdL:GDP:Mn soaked with GDP-Glc | | Descriptor: | GDP-ALPHA-D-GLUCOSE, MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPS
 
 | | Crystal structrue of MtdL:GDP:Mn | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7XPU
 
 | | crystal structure of MtdL-S228A-His soaked GDP-Fucp and Mn | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, MANGANESE (II) ION, ... | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
3N40
 
 | | Crystal structure of the immature envelope glycoprotein complex of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N42
 
 | | Crystal structures of the mature envelope glycoprotein complex (furin cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
