1X1L
 
 | | Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit. | | Descriptor: | GTP-binding protein era, RNA (130-MER) | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
5T8L
 
 | |
5T8N
 
 | |
6LYW
 
 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
8IS0
 
 | | Carbon Sulfoxide lyase - Y106F | | Descriptor: | 2-AMINO-ACRYLIC ACID, PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRK
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRY
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8IRZ
 
 | | Carbon Sulfoxide lyase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Probable hercynylcysteine sulfoxide lyase | | Authors: | Gong, W.M, Wei, L.L, Liu, L. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of mycobacterial ergothioneine-biosynthesis C-S lyase EgtE.
J.Biol.Chem., 300, 2024
|
|
8JZV
 
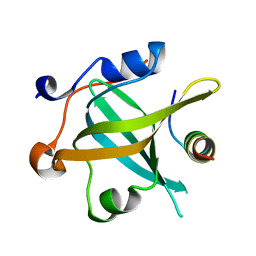 | | RPA70N-ETAA1 fusion | | Descriptor: | Ewing's tumor-associated antigen 1, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8JZY
 
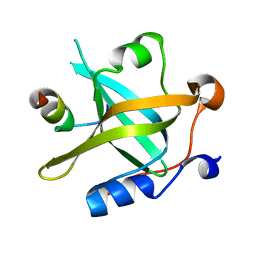 | | RPA70N-RAD9 fusion | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8K00
 
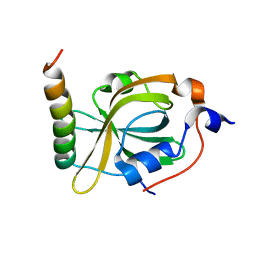 | | RPA70N-MRE11 fusion | | Descriptor: | Double-strand break repair protein MRE11, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
2R0Q
 
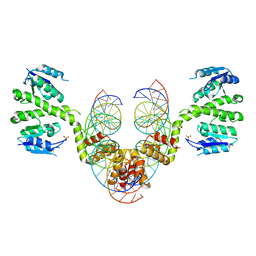 | |
8K1Y
 
 | | Crystal structure of human serum albumin and ruthenium Val complex adduct | | Descriptor: | Albumin, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gong, W.J, Xie, L.L, Wang, W.M, Wang, H.F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the anti-proliferation activity and photoinduced NO release of four nitrosylruthenium isomeric complexes and their HSA complex adducts.
Metallomics, 16, 2024
|
|
2PIJ
 
 | | Structure of the Cro protein from prophage Pfl 6 in Pseudomonas fluorescens Pf-5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, Prophage Pfl 6 Cro, ... | | Authors: | Roessler, C.G, Roberts, S.A, Montfort, W.R, Cordes, M.H.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1R9H
 
 | |
1R35
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER, TETRAHYDROBIOPTERIN AND 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE | | Descriptor: | 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Shieh, H.S, Stevens, A.M, Stallings, W.C. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4-Fluorinated L-lysine analogs as selective i-NOS inhibitors: methodology for introducing fluorine into the lysine side chain.
Org.Biomol.Chem., 1, 2003
|
|
1SAK
 
 | |
1SAE
 
 | |
1SAF
 
 | |
1SAL
 
 | |
1SOF
 
 | | Crystal structure of the azotobacter vinelandii bacterioferritin at 2.6 A resolution | | Descriptor: | BARIUM ION, Bacterioferritin, FE (II) ION, ... | | Authors: | Liu, H.L, Huang, J.F, Bi, R.C. | | Deposit date: | 2004-03-14 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A resolution crystal structure of the bacterioferritin from Azotobacter vinelandii
Febs Lett., 573, 2004
|
|
6U4T
 
 | | Carbonic anhydrase 9 in complex with SB4197 | | Descriptor: | 4-methyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-08-26 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
6UFD
 
 | |
6UGQ
 
 | | Human Carbonic Anhydrase IX-mimic complexed with SB4-206 | | Descriptor: | 5-chloro-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase IX-mimic, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
6UH0
 
 | | Human Carbonic Anhydrase 2 in complex with SB4-202 | | Descriptor: | 4-methyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Lomelino, C.L, McKenna, R. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.306 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
