4NU7
 
 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | Descriptor: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NV4
 
 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
4NMW
 
 | | Crystal Structure of Carboxylesterase BioH from Salmonella enterica | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pimelyl-[acyl-carrier protein] methyl ester esterase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Crystal Structure of Carboxylesterase BioH from Salmonella enterica
To be Published
|
|
4NOH
 
 | | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Lipoprotein, putative | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4NU9
 
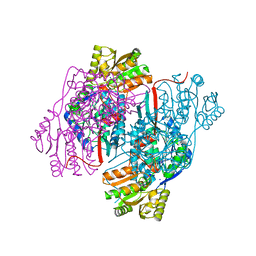 | | 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Stam, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OVD
 
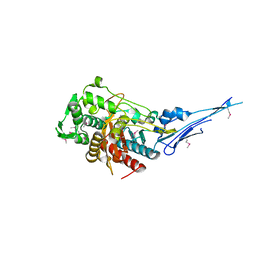 | | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469 | | Descriptor: | CALCIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Babnigg, G, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469
To be Published
|
|
4OHJ
 
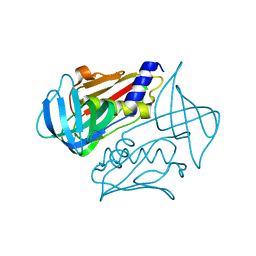 | | Crystal structure of toxic shock syndrome toxin-1 (TSST-1) from Staphylococcus aureus | | Descriptor: | Toxic shock syndrome toxin-1 | | Authors: | Filippova, E.V, Shuvalova, L, Flores, K, Dubrovska, I, Bishop, B, Ngo, H, Yeats, C, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of toxic shock syndrome toxin-1 (TSST-1) from Staphylococcus aureus
To be Published
|
|
4OEN
 
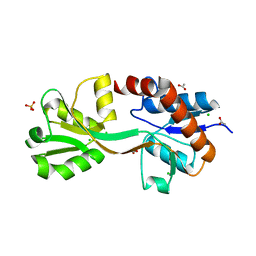 | | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | ACETATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second substrate binding domain of a putative amino acid ABC transporter from Streptococcus pneumoniae Canada MDR_19A
To be Published
|
|
4OC9
 
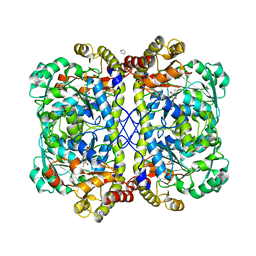 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
4ONX
 
 | | 2.8 Angstrom Crystal Structure of Sensor Domain of Histidine Kinase from Clostridium perfringens. | | Descriptor: | CHLORIDE ION, SULFATE ION, Sensor histidine kinase, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of Sensor Domain of Histidine Kinase from Clostridium perfringens.
TO BE PUBLISHED
|
|
4NHD
 
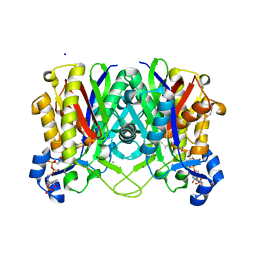 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
4NMY
 
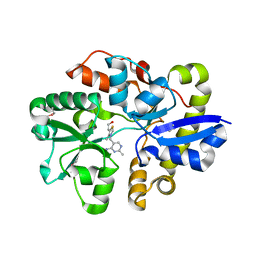 | | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ABC-type transport system, extracellular solute-binding protein | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile
To be Published
|
|
4NOI
 
 | | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase subunit alpha, IODIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4NP6
 
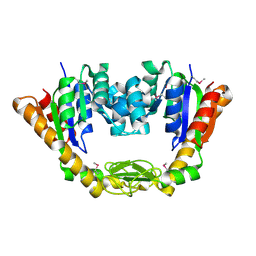 | | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor | | Descriptor: | Adenylate kinase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor
To be Published
|
|
4NLM
 
 | | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo1340 protein | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e
To be Published
|
|
4NF2
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | CHLORIDE ION, NORVALINE, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Handing, K, Cymborowski, M.T, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4NEL
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine | | Descriptor: | N,N-dimethylmethanamine, Transcriptional regulator | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with N,N-dimethylmethanamine
To be Published
|
|
4O0N
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NZP
 
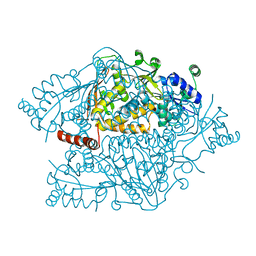 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4NCZ
 
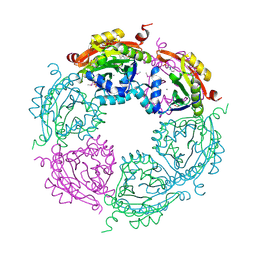 | | Spermidine N-acetyltransferase from Vibrio cholerae in complex with 2-[n-cyclohexylamino]ethane sulfonate. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, SODIUM ION, ... | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4NEG
 
 | | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | FORMIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4MY1
 
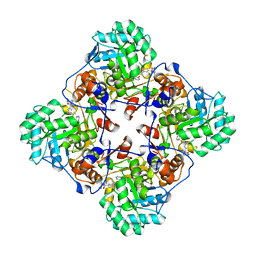 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | Descriptor: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4M9D
 
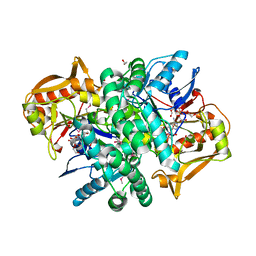 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
4MVJ
 
 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | Descriptor: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
