5IOB
 
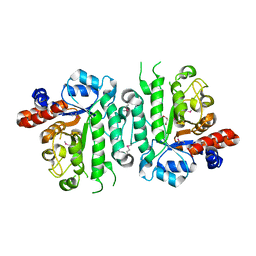 | | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase-related glycosidases, CHLORIDE ION, ... | | Authors: | Chang, C, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum
To Be Published
|
|
1K4N
 
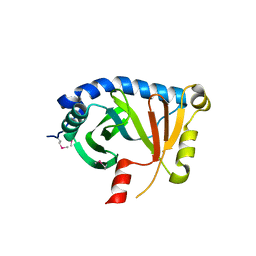 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
2ZC2
 
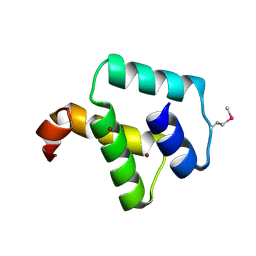 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
3GE2
 
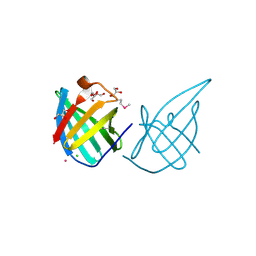 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3ODF
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Second step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ODD
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Second step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3B7H
 
 | |
3B4S
 
 | |
3B49
 
 | |
3B85
 
 | |
3B4Q
 
 | |
3B79
 
 | |
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3BEE
 
 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Putative YfrE protein | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
3BHG
 
 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
3BIO
 
 | | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83.
To be Published
|
|
3BJ6
 
 | | Crystal structure of MarR family transcription regulator SP03579 | | Descriptor: | ETHANOL, Transcriptional regulator, MarR family | | Authors: | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of MarR family Transcription Regulator SP03579.
To be Published
|
|
3BJO
 
 | |
3B73
 
 | |
3B8B
 
 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
3U1B
 
 | | Crystal structure of the S238R mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin immunity protein MccF | | Authors: | Nocek, B, Gu, M, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
3SFP
 
 | | Crystal Structure of the Mono-Zinc-boundform of New Delhi Metallo-beta-Lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
