7XBZ
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with R-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-[(1R)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7X70
 
 | |
7X6Y
 
 | |
7WO9
 
 | | Cryo-EM structure of full-length Nup188 | | Descriptor: | Nucleoporin NUP188 | | Authors: | Zhao, L, Li, Z.Q, Sui, S.F. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7X6S
 
 | |
7X6V
 
 | |
7XH8
 
 | | The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike | | Descriptor: | Spike glycoprotein, The heavy chain of ZCB11 antibody, The light chain of ZCB11 antibody | | Authors: | Hang, L, Dang, S. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge.
Nat Commun, 13, 2022
|
|
7XVG
 
 | | Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35 | | Descriptor: | AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7BZO
 
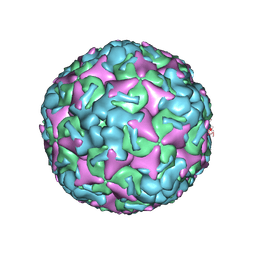 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Z
 
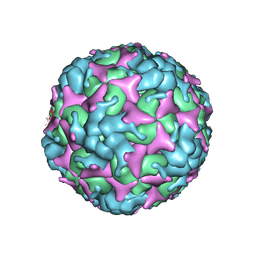 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4T
 
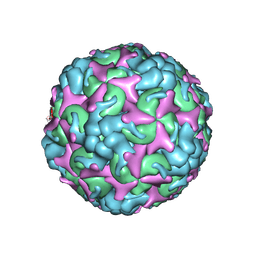 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4W
 
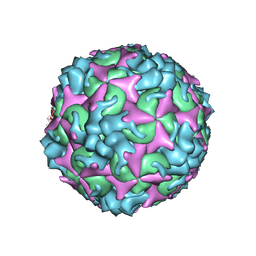 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DLX
 
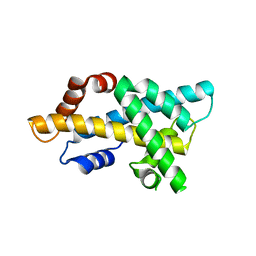 | | crystal structure of H2AM4>Z-H2B | | Descriptor: | Histone H2B,Histone H2A | | Authors: | Dai, L.C, Zhou, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange.
Cell Rep, 35, 2021
|
|
7DRQ
 
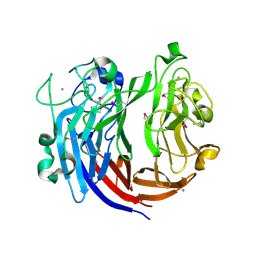 | | Crystal structure of polysaccharide lyase Uly1 | | Descriptor: | CALCIUM ION, Uly1 | | Authors: | Chen, X.L, Cao, H.Y, Xu, F, Dong, F. | | Deposit date: | 2020-12-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanistic Insights into Substrate Recognition and Catalysis of a New Ulvan Lyase of Polysaccharide Lyase Family 24.
Appl.Environ.Microbiol., 87, 2021
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZN
 
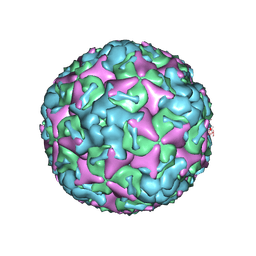 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CZH
 
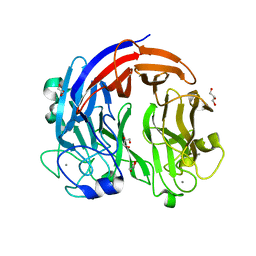 | | PL24 ulvan lyase-Uly1 | | Descriptor: | CALCIUM ION, GLYCEROL, Uly1 | | Authors: | Zhang, Y.Z, Chen, X.L, Dong, F, Xu, F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Mechanistic Insights into Substrate Recognition and Catalysis of a New Ulvan Lyase of Polysaccharide Lyase Family 24.
Appl.Environ.Microbiol., 87, 2021
|
|
7DFT
 
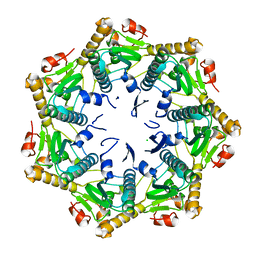 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
7DFU
 
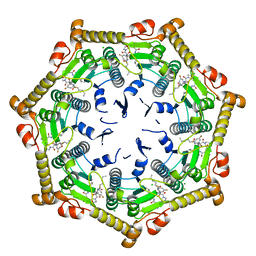 | |
7BZU
 
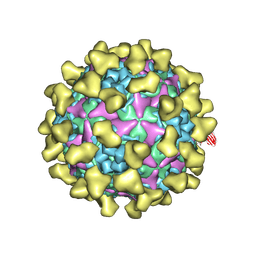 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Y
 
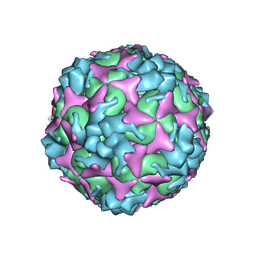 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U0D
 
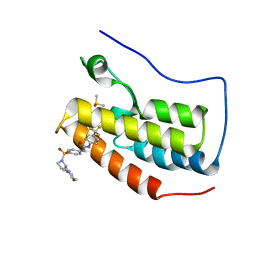 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
6UWY
 
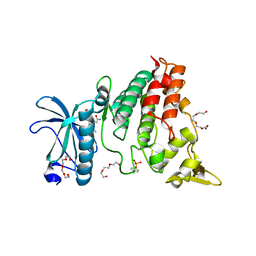 | | DYRK1A bound to a harmine derivative | | Descriptor: | 4-(7-methoxy-1-methyl-9H-beta-carbolin-9-yl)butanamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, TETRAETHYLENE GLYCOL | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-11-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and Biological Validation of a Harmine-Based, Central Nervous System (CNS)-Avoidant, Selective, Human beta-Cell Regenerative Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase A (DYRK1A) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6UWU
 
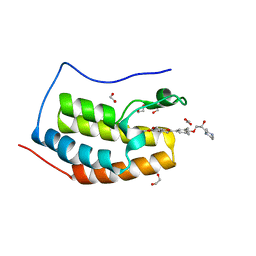 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
3F5C
 
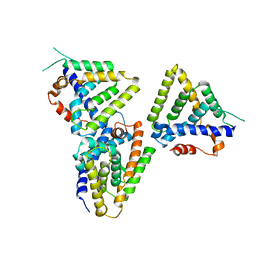 | | Structure of Dax-1:LRH-1 complex | | Descriptor: | Nuclear receptor subfamily 0 group B member 1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Fletterick, R.J, Sablin, E.P. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
