3K4W
 
 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
1SV6
 
 | | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli | | Descriptor: | 2-keto-4-pentenoate hydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sharp, A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-03-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli
To be Published
|
|
3SLR
 
 | | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis containing phosphatase domain complexed with Mg. | | Descriptor: | MAGNESIUM ION, uncharacterized protein BF1531 | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis
containing phosphatase domain complexed with Mg.
TO BE PUBLISHED
|
|
3OXN
 
 | |
2OX4
 
 | | Crystal structure of putative dehydratase from Zymomonas mobilis ZM4 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Zymomonas Mobilis Zm4
To be Published
|
|
2OZ3
 
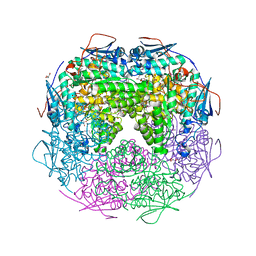 | | Crystal structure of L-Rhamnonate dehydratase from Azotobacter vinelandii | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Freeman, J.C, Bain, K, Gheyi, T, Wu, B, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Rhamnonate dehydratase from azotobacter vinelandii
To be Published
|
|
1VQV
 
 | |
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | Descriptor: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
2POD
 
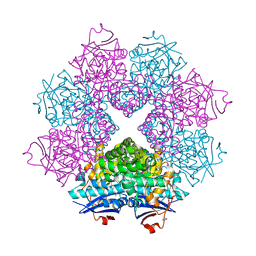 | | Crystal structure of a member of enolase superfamily from Burkholderia pseudomallei K96243 | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Ozyurt, S, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-06-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Burkholderia Pseudomallei.
To be Published
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
3GAZ
 
 | |
3GBW
 
 | | Crystal structure of the first PHR domain of the Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Klemke, R.L, Miller, S.A, Bain, K.T, Rutter, M.E, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
1SGM
 
 | |
3GL3
 
 | |
3GV0
 
 | |
3EOI
 
 | | CRYSTAL STRUCTURE OF putative PROTEIN PilM from Escherichia coli B7A | | Descriptor: | PilM | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J.B, Sauder, J.M, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3EEY
 
 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
3H7V
 
 | | CRYSTAL STRUCTURE OF O-SUCCINYLBENZOATE SYNTHASE FROM THERMOSYNECHOCOCCUS ELONGATUS BP-1 complexed with MG in the active site | | Descriptor: | MAGNESIUM ION, O-SUCCINYLBENZOATE SYNTHASE | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3H70
 
 | | Crystal structure of o-succinylbenzoic acid synthetase from staphylococcus aureus Complexed with mg in the active site | | Descriptor: | MAGNESIUM ION, O-succinylbenzoic acid (OSB) synthetase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3HG7
 
 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
3HIN
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Putative 3-hydroxybutyryl-CoA dehydratase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009
To be Published
|
|
3PU5
 
 | |
1RZN
 
 | |
1TK9
 
 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
1U6L
 
 | |
