8R6Z
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Apo form 2 | | Descriptor: | Heparinase, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R73
 
 | | Polysaccharide lyase BcPL33HA (BcellWH2_04512) Apo form | | Descriptor: | Heparinase II/III-like protein, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R75
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Apo form 1 | | Descriptor: | Heparinase, ZINC ION | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R72
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HA dp4 collected at 1.33 A | | Descriptor: | Heparinase, ZINC ION, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R70
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HA dp4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8R71
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HAdp4 collected at 1.22 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ZINC ION, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7UR7
 
 | |
7UR8
 
 | |
6XNS
 
 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XI6
 
 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XH5
 
 | |
6XT4
 
 | |
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
5LP2
 
 | |
2UVK
 
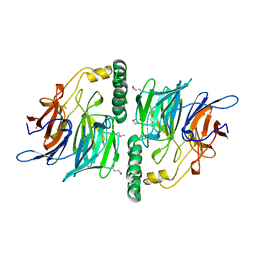 | | Structure of YjhT | | Descriptor: | YJHT | | Authors: | Muller, A, Severi, E, Wilson, K.S, Thomas, G.H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sialic Acid Mutarotation is Catalyzed by the Escherichia Coli Beta-Propeller Protein Yjht.
J.Biol.Chem., 283, 2008
|
|
3ZRZ
 
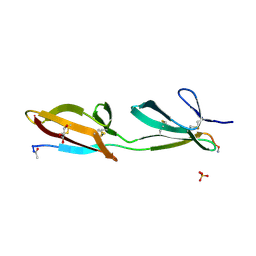 | | Crystal structure of the second and third fibronectin F1 modules in complex with a fragment of Streptococcus pyogenes SfbI-5 | | Descriptor: | FIBRONECTIN, FIBRONECTIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Norris, N.C, Bingham, R.J, Potts, J.R. | | Deposit date: | 2011-06-21 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analysis of the Tandem Beta-Zipper Interaction of a Streptococcal Protein with Human Fibronectin.
J.Biol.Chem., 286, 2011
|
|
4WMV
 
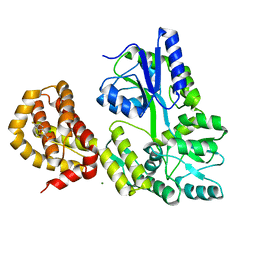 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | Descriptor: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Moulin, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6YCH
 
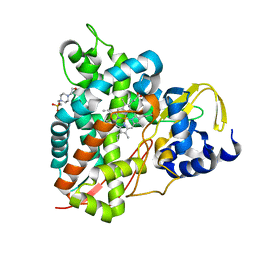 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
2CMP
 
 | | crystal structure of the DNA binding domain of G1P SMALL TERMINASE SUBUNIT from bacteriophage SF6 | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Benini, S, Chechik, M, Ortiz-Lombardia, M, Polier, S, Shevtsov, M.B, DeLuchi, D, Alonso, J.C, Antson, A.A. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The 1.58 A Resolution Structure of the DNA-Binding Domain of Bacteriophage Sf6 Small Terminase Provides New Hints on DNA Binding
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6J6L
 
 | |
3LD0
 
 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
3LCZ
 
 | | B.licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
