6BI8
 
 | |
4Z4S
 
 | | Crystal structure of GII.10 P domain in complex with 150mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4Z4V
 
 | | Crystal structure of GII.10 P domain in complex with 19mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, NITRATE ION, ... | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
6BIB
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
1BDN
 
 | |
4BWZ
 
 | |
4YPT
 
 | | X-ray structural of three tandemly linked domains of nsp3 from murine hepatitis virus at 2.60 Angstroms resolution | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Chen, Y, Savinov, S.N, Mielech, A.M, Cao, T, Baker, S.C, Mesecar, A.D. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | X-ray Structural and Functional Studies of the Three Tandemly Linked Domains of Non-structural Protein 3 (nsp3) from Murine Hepatitis Virus Reveal Conserved Functions.
J.Biol.Chem., 290, 2015
|
|
4C0S
 
 | | Mammalian translation elongation factor eEF1A2 | | Descriptor: | ELONGATION FACTOR 1-ALPHA 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Crepin, T, Shalak, V.F, Yaremchuk, A.D, Vlasenko, D.O, McCarthy, A.A, Negrutskii, B.S, Tukalo, M.A, El'skaya, A.V. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-27 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Mammalian Translation Elongation Factor Eef1A2: X-Ray Structure and New Features of Gdp/GTP Exchange Mechanism in Higher Eukaryotes
Nucleic Acids Res., 42, 2014
|
|
4WEL
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
4ERJ
 
 | |
6B0X
 
 | |
6B3E
 
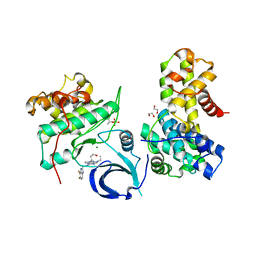 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
6B23
 
 | |
4Z4Z
 
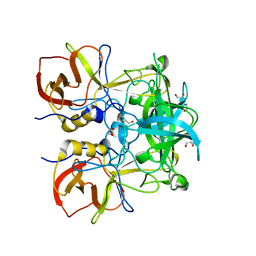 | | Crystal structure of GII.10 P domain in complex with 30mM B antigen (trisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4YZF
 
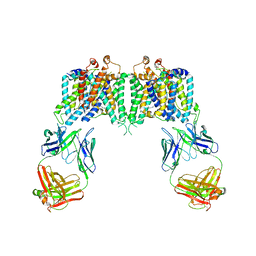 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
4Z4U
 
 | | Crystal structure of GII.10 P domain in complex with 37.5mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
1B8Y
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXED WITH NON-PEPTIDE INHIBITORS: IMPLICATIONS FOR INHIBITOR SELECTIVITY | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), SULFATE ION, ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-03 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
4DTK
 
 | | Novel and selective pan-PIM kinase inhibitor | | Descriptor: | (5Z)-5-{2-[(3R)-3-aminopiperidin-1-yl]-3-(propan-2-yloxy)benzylidene}-1,3-thiazolidine-2,4-dione, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2012-02-21 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of novel benzylidene-1,3-thiazolidine-2,4-diones as potent and selective inhibitors of the PIM-1, PIM-2, and PIM-3 protein kinases.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
4D1C
 
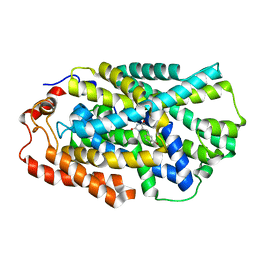 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1BWS
 
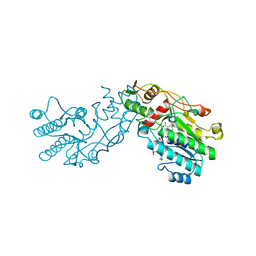 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | Authors: | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
1BXK
 
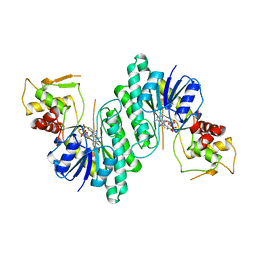 | | DTDP-GLUCOSE 4,6-DEHYDRATASE FROM E. COLI | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) | | Authors: | Thoden, J.B, Hegeman, A.D, Frey, P.A, Holden, H.M. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Structure of Dtdp-Glucose 4,6-Dehydratase from E. Coli
Protein Sci.
|
|
