5ZRZ
 
 | | Crystal Structure of EphA5/SAMD5 Complex | | Descriptor: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
4ELX
 
 | | Structure of apo E.coli. 1,4-dihydroxy-2- naphthoyl CoA synthases with Cl | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
5DQU
 
 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
3DC1
 
 | | Crystal structure of kynurenine aminotransferase II complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Kynurenine/alpha-aminoadipate aminotransferase mitochondrial | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Biosci.Rep., 28, 2008
|
|
5DQT
 
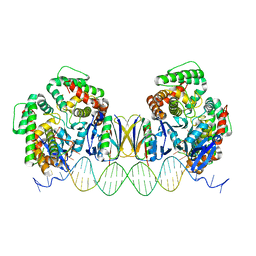 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5DQZ
 
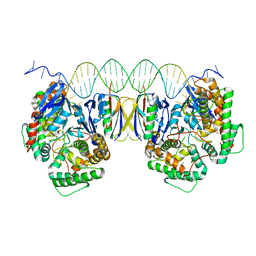 | | Crystal Structure of Cas-DNA-PAM complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (36-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
8VJB
 
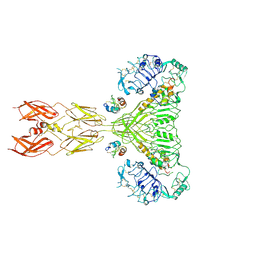 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJC
 
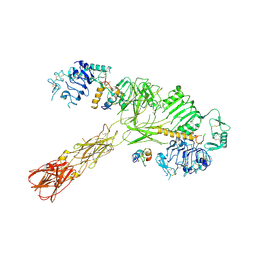 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
7C23
 
 | | Crystal structure of CrmE10, a SGNH-hydrolase family esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
8Y37
 
 | | Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
8Y38
 
 | | Cryo-EM structure of Staphylococcus aureus 70S ribosome (strain 15B196) in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
8Y36
 
 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
5EJO
 
 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
3EXF
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
8YKJ
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with X77 | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, Non-structural protein 11 | | Authors: | Zhou, X.L, Lin, C, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
8YKO
 
 | |
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXE
 
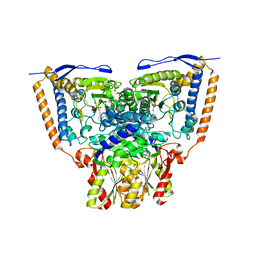 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
8Y39
 
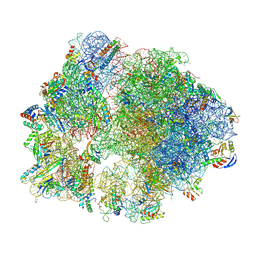 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
3EXI
 
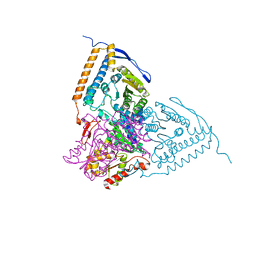 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex with the subunit-binding domain (SBD) of E2p, but SBD cannot be modeled into the electron density | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
1W99
 
 | |
3EXH
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
5DLJ
 
 | | Crystal Structure of Cas-DNA-N1 complex | | Descriptor: | 39-mer DNA N1-F, 39-mer DNA N1-R, CRISPR-associated endonuclease Cas1, ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-05 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
7C29
 
 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
