7XE0
 
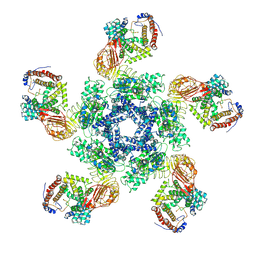 | | Cryo-EM structure of plant NLR Sr35 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8I
 
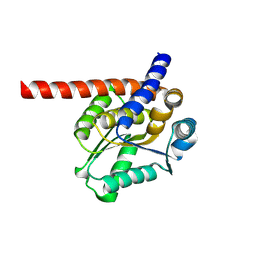 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
6BWU
 
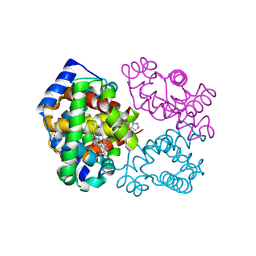 | | Crystal structure of carboxyhemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
7E3O
 
 | |
6BWP
 
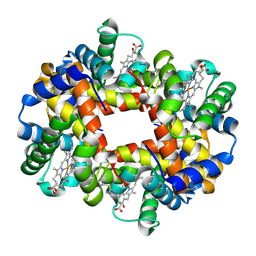 | | Crystal structure of Deoxy Hemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
8K8F
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7Z
 
 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7M
 
 | | De novo design protein -T01 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K83
 
 | | De novo design protein -N2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, De novo design protein, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K84
 
 | | De novo design protein -N3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KAC
 
 | | De novo design protein -NX1 | | Descriptor: | De novo design protein -NX1 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8G
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC4
 
 | | De novo design protein -NA05 | | Descriptor: | De novo design protein -NA05 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KDQ
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein -T03 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC8
 
 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCJ
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA7
 
 | | De novo design protein -NB7 | | Descriptor: | De novo design protein -NB7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KA6
 
 | | De novo design protein -NA7 | | Descriptor: | De novo design protein -NA7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KCK
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
6O5G
 
 | | Calmodulin in complex with isomalbrancheamide D | | Descriptor: | (5aS,12aS,13aS)-9-bromo-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7 ,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2019-03-02 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EEB
 
 | | Calmodulin in complex with malbrancheamide | | Descriptor: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
