4EE0
 
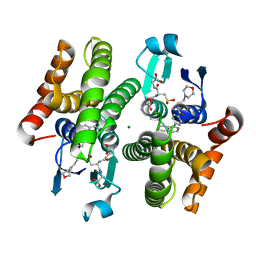 | | Crystal structure of hH-PGDS with water displacing inhibitor | | Descriptor: | 4-(isoquinolin-1-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, Hematopoietic prostaglandin D synthase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, ... | | Authors: | Day, J.E, Thorarensen, A, Trujillo, J.I. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Investigation of the binding pocket of human hematopoietic prostaglandin (PG) D2 synthase (hH-PGDS): a tale of two waters.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7MM0
 
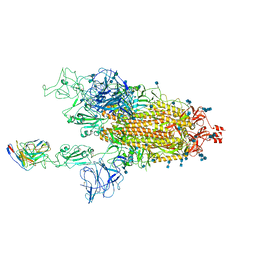 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
3T56
 
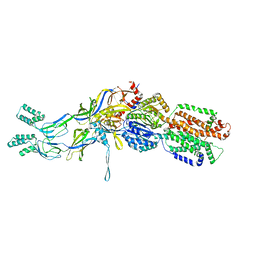 | | Crystal structure of the pre-extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
3TFQ
 
 | | Crystal structure of 11b-hsd1 double mutant (l262r, f278e) complexed with 8-{[(2-CYANOPYRIDIN-3-YL)METHYL]SULFANYL}-6-HYDROXY-3,4-DIHYDRO-1H-PYRANO[3,4-C]PYRIDINE-5-CARBONITRILE | | Descriptor: | 8-{[(2-cyanopyridin-3-yl)methyl]sulfanyl}-6-hydroxy-3,4-dihydro-1H-pyrano[3,4-c]pyridine-5-carbonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 3-hydroxy-4-cyano-isoquinolines as novel, potent, and selective inhibitors of human 11beta-hydroxydehydrogenase 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3T99
 
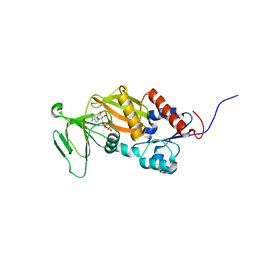 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and in the absence of cadmium at pH 7.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
4ER5
 
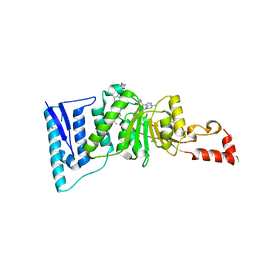 | | Crystal structure of human DOT1L in complex with 2 molecules of EPZ004777 | | Descriptor: | 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
7M7D
 
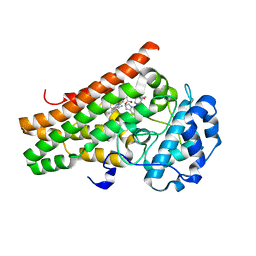 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
3TGR
 
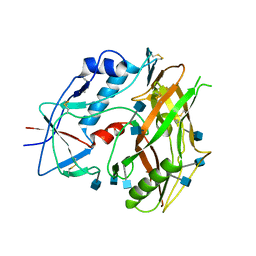 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7MPS
 
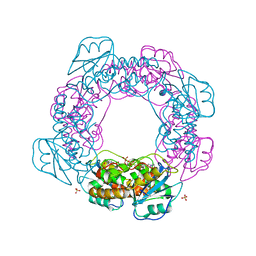 | | Brucella melitensis NrnC with engaged loop | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7M63
 
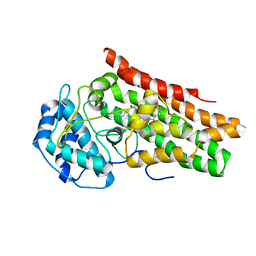 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-70099 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[(1R,3S,5S,6r)-3-(5,6-difluoro-1H-benzimidazol-1-yl)bicyclo[3.1.0]hexan-6-yl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
7MQD
 
 | | Bartonella henselae NrnC complexed with pAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MTD
 
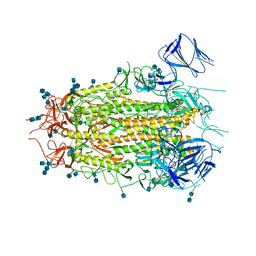 | | Structure of aged SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MPT
 
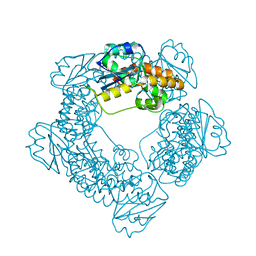 | | Brucella melitensis NrnC with bound Mg2+ | | Descriptor: | MAGNESIUM ION, NanoRNase C, PHOSPHATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQF
 
 | | Bartonella henselae NrnC complexed with pAAAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPU
 
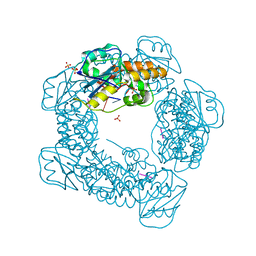 | | Brucella melitensis NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPP
 
 | |
4E4M
 
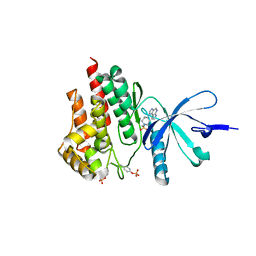 | | JAK2 kinase (JH1 domain) in complex with compound 30 | | Descriptor: | 1-[4-methyl-1-(methylsulfonyl)piperidin-4-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
7MQE
 
 | | Bartonella henselae NrnC complexed with pAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQB
 
 | | Bartonella henselae NrnC bound to pGG. D4 Symmetry | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MTC
 
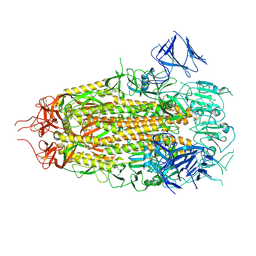 | | Structure of freshly purified SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MPR
 
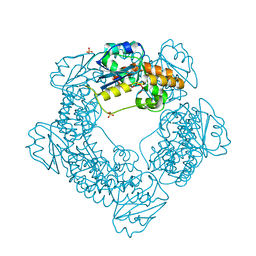 | | Brucella melitensis NrnC | | Descriptor: | NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQC
 
 | | Bartonella henselae NrnC bound to pGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPO
 
 | | Bartonella henselae NrnC bound to pAp | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPL
 
 | | Bartonella henselae NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MTE
 
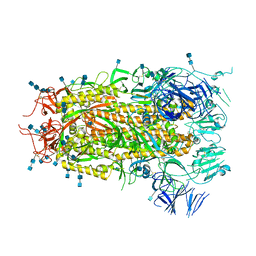 | |
