1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
3ST7
 
 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Kuroda, M, Yao, M, Watanabe, M, Ohta, T, Tanaka, I, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-07-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
3OED
 
 | |
3VHR
 
 | | Crystal Structure of capsular polysaccharide assembling protein CapF from Staphylococcus aureus in space group C2221 | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, FORMIC ACID, ZINC ION | | Authors: | Miyafusa, T, Yoshikazu, T, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains.
Biochem.J., 443, 2012
|
|
5MN4
 
 | | S. aureus FtsZ 12-316 F138A GDP Open form (1FOf) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN5
 
 | | S. aureus FtsZ 12-316 T66W GTP Closed form (2TCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN6
 
 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
6TGG
 
 | | scFv-1SM3 in complex with glycopeptide containing an sp2-imino sugar | | Descriptor: | 1,2-ETHANEDIOL, Mucin-1, ScFv_SM3, ... | | Authors: | Bermejo, I.A, Navo, C.D, Castro-Lopez, J, Samchez-Fernandez, E.M, Guerreiro, A, Avenoza, A, Busto, J.H, Garcia-Martin, F, Nishimura, S.I, Garcia-Fernandez, J.M, Ortiz-Mellet, C, Bernardes, G.J.L, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Chem Sci, 11, 2020
|
|
5ND1
 
 | | Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus | | Descriptor: | Capsid protein | | Authors: | Mata, C.P, Luque, D, Gomez Blanco, J, Rodriguez, J.M, Suzuki, N, Ghabrial, S.A, Carrascosa, J.L, Trus, B.L, Caston, J.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.
PLoS Pathog., 13, 2017
|
|
5N8Y
 
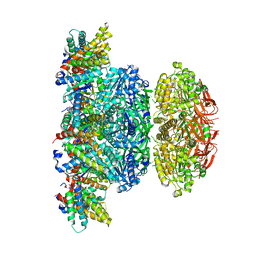 | | KaiCBA circadian clock backbone model based on a Cryo-EM density | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Schuller, J.M, Snijder, J, Loessl, P, Heck, A.J.R, Foerster, F. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state.
Science, 355, 2017
|
|
5MN8
 
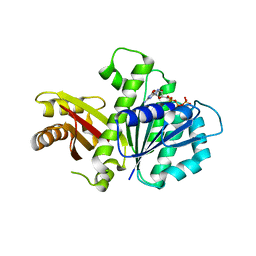 | | S. aureus FtsZ 12-316 F138A GTP Closed form (5FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN7
 
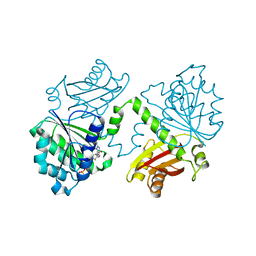 | | S. aureus FtsZ 12-316 F138A GTP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
2MN3
 
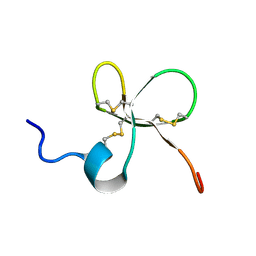 | | Structure of Platypus 'Intermediate' Defensin-like Peptide (Int-DLP) | | Descriptor: | Defensin-BvL | | Authors: | Torres, A.M, Bansal, P.S, Koh, J.M.S, Pages, G, Wu, M.J, Kuchel, P.W. | | Deposit date: | 2014-03-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and antimicrobial activity of platypus 'intermediate' defensin-like peptide.
Febs Lett., 588, 2014
|
|
2MPA
 
 | | BACTERICIDAL ANTIBODY AGAINST NEISSERIA MENINGITIDIS | | Descriptor: | CADMIUM ION, CONJUGATE OF PORA P1.16 PEPTIDE WITH FLUORESCEIN, MN12H2 IGG2A-KAPPA, ... | | Authors: | Van Den Elsen, J.M.H, Herron, J.N, Kroon, J, Gros, P. | | Deposit date: | 1999-06-09 | | Release date: | 1999-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bactericidal antibody recognition of a PorA epitope of Neisseria meningitidis: crystal structure of a Fab fragment in complex with a fluorescein-conjugated peptide.
Proteins, 29, 1997
|
|
1HXS
 
 | | CRYSTAL STRUCTURE OF MAHONEY STRAIN OF POLIOVIRUS AT 2.2A RESOLUTION | | Descriptor: | GENOME POLYPROTEIN, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | Authors: | Miller, S.T, Hogle, J.M, Filman, D.J. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
5EZT
 
 | | Peracetylated Bovine Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Whitesides, G.M, Kang, K, Choi, J.-M, Fox, J.M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-07-20 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Acetylation of Surface Lysine Groups of a Protein Alters the Organization and Composition of Its Crystal Contacts.
J.Phys.Chem.B, 120, 2016
|
|
5DS3
 
 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
5DSY
 
 | | Crystal structure of constitutively active PARP-2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Riccio, A.A, Pascal, J.M. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
6ZQK
 
 | | HER2-binding scFv-Fab fusion 841 | | Descriptor: | 1,2-ETHANEDIOL, 841 heavy chain, 841 light chain | | Authors: | Kast, F, Schwill, M, Stueber, J.C, Pfundstein, S, Nagy-Davidescu, G, Monne Rodriguez, J.M, Seehusen, F, Richter, C.P, Honegger, A, Hartmann, K.P, Weber, T.G, Kroener, F, Ernst, P, Piehler, J, Plueckthun, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action.
Nat Commun, 12, 2021
|
|
8D5K
 
 | |
8D5E
 
 | | The complex of Gtf2b Peptide TGAASFDEF Presented by H2-Dd | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Custodio, J.M.F, Baker, B.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and physical features that distinguish tumor-controlling from inactive cancer neoepitopes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D5F
 
 | |
7OI2
 
 | | SaFtsz complexed with GDP (NaCl purification) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu Morales, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
2HUO
 
 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JOL
 
 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
