1U24
 
 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U25
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate in the C2221 crystal form | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U26
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
3A8T
 
 | | Plant adenylate isopentenyltransferase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylate isopentenyltransferase, PHOSPHATE ION | | Authors: | Chu, H.-M, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-10-09 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure and substrate specificity of plant adenylate isopentenyltransferase from Humulus lupulus: distinctive binding affinity for purine and pyrimidine nucleotides
Nucleic Acids Res., 38, 2010
|
|
3AQ0
 
 | | Ligand-bound form of Arabidopsis medium/long-chain length prenyl pyrophosphate synthase (surface polar residue mutant) | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, DI(HYDROXYETHYL)ETHER, FARNESYL DIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase
Plant Physiol., 155, 2011
|
|
3APZ
 
 | |
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
1D39
 
 | | COVALENT MODIFICATION OF GUANINE BASES IN DOUBLE STRANDED DNA: THE 1.2 ANGSTROMS Z-DNA STRUCTURE OF D(CGCGCG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(CU)GP*CP*(CU)GP*CP*(CU)G)-3'), SODIUM ION | | Authors: | Kagawa, T.F, Geierstanger, B.H, Wang, A.H.-J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Covalent modification of guanine bases in double-stranded DNA. The 1.2-A Z-DNA structure of d(CGCGCG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
1D35
 
 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(A40)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D36
 
 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1DCG
 
 | | THE MOLECULAR STRUCTURE OF THE LEFT-HANDED Z-DNA DOUBLE HELIX AT 1.0 ANGSTROM ATOMIC RESOLUTION. GEOMETRY, CONFORMATION, AND IONIC INTERACTIONS OF D(CGCGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gessner, R.V, Frederick, C.A, Quigley, G.J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG).
J.Biol.Chem., 264, 1989
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
1NDN
 
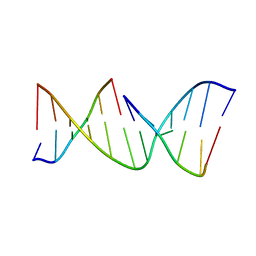 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1DSD
 
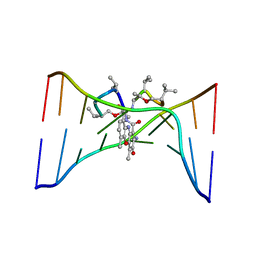 | |
1DSC
 
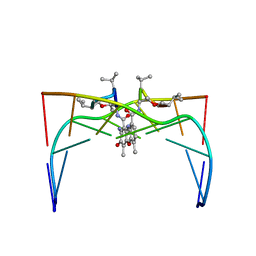 | |
3MBR
 
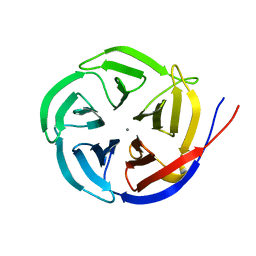 | | Crystal Structure of the Glutaminyl Cyclase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Glutamine cyclotransferase | | Authors: | Huang, W.-L, Wang, Y.-R, Ko, T.-P, Chia, C.-Y, Huang, K.-F, Wang, A.H.-J. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure and functional analysis of the glutaminyl cyclase from Xanthomonas campestris
J.Mol.Biol., 401, 2010
|
|
1MNV
 
 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
3RNZ
 
 | |
3RO1
 
 | |
3RO0
 
 | |
5G50
 
 | |
3DR2
 
 | | Structural and Functional Analyses of XC5397 from Xanthomonas campestris: A Gluconolactonase Important in Glucose Secondary Metabolic Pathways | | Descriptor: | CALCIUM ION, Exported gluconolactonase | | Authors: | Chen, C.-N, Chin, K.-H, Wang, A.H.-J, Chou, S.H. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The First Crystal Structure of Gluconolactonase Important in the Glucose Secondary Metabolic Pathways
J.Mol.Biol., 384, 2008
|
|
3EK5
 
 | | Unique GTP-binding Pocket and Allostery of UMP Kinase from a Gram-Negative Phytopathogen Bacterium | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Uridylate kinase | | Authors: | Tu, J.-L, Chin, K.-H, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Unique GTP-Binding Pocket and Allostery of Uridylate Kinase from a Gram-Negative Phytopathogenic Bacterium
J.Mol.Biol., 385, 2009
|
|
3EK6
 
 | |
