3OTJ
 
 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8IHW
 
 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHY
 
 | | X-ray crystal structure of Q387E mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
6M4L
 
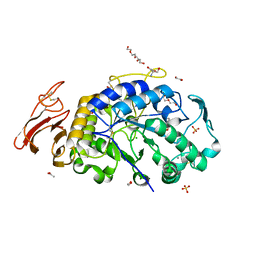 | | X-ray crystal structure of the E249Q mutant of alpha-amylase I from Eisenia fetida | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8K9N
 
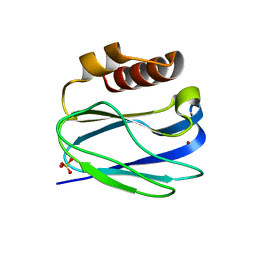 | | Subatomic resolution structure of Pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Lintuluoto, M, Kurihara, K, Hasegawa, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
8K9P
 
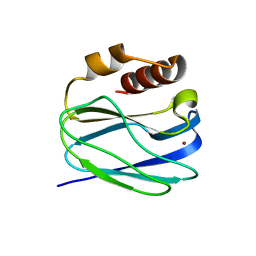 | | Neutron X-ray joint structure of pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Kurihara, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
6L46
 
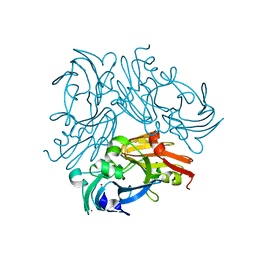 | | High-resolution neutron and X-ray joint refined structure of copper-containing nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.3 Å), X-RAY DIFFRACTION | | Cite: | High-resolution neutron crystallography visualizes an OH-bound resting state of a copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M3D
 
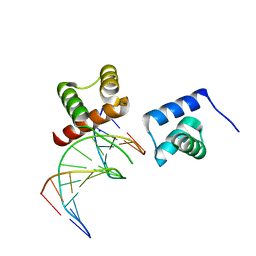 | | X-ray crystal structure of tandemly connected engrailed homeodomains (EHD) with R53A mutations and DNA complex | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*TP*AP*GP*GP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*TP*AP*AP*TP*CP*C)-3'), SODIUM ION, ... | | Authors: | Sunami, T, Hirano, Y, Tamada, T, Kono, H. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for designing an array of engrailed homeodomains.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4REN
 
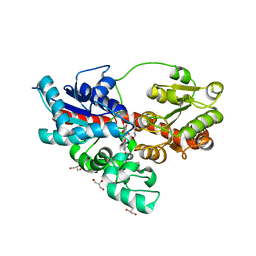 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
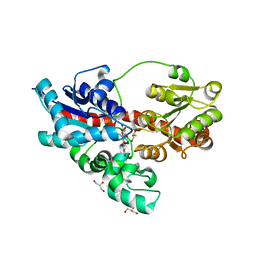 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
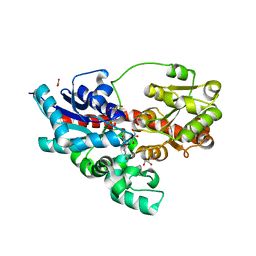 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
1EHA
 
 | | CRYSTAL STRUCTURE OF GLYCOSYLTREHALOSE TREHALOHYDROLASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1EH9
 
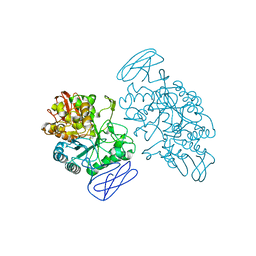 | | CRYSTAL STRUCTURE OF SULFOLOBUS SOLFATARICUS GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
3FX5
 
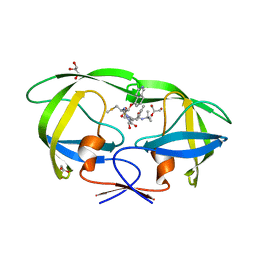 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
6M4K
 
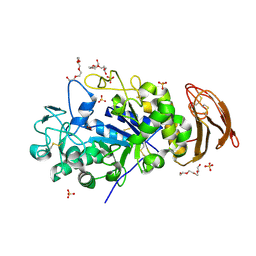 | | X-ray crystal structure of wild type alpha-amylase I from Eisenia fetida | | Descriptor: | ACETATE ION, Alpha-amylase, CALCIUM ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M4M
 
 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | Descriptor: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6IQE
 
 | | Human prohibitin 2 | | Descriptor: | Prohibitin-2 | | Authors: | Hirano, Y, Koshiba, T, Tamada, T. | | Deposit date: | 2018-11-07 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural Basis of Mitochondrial Scaffolds by Prohibitin Complexes: Insight into a Role of the Coiled-Coil Region.
Iscience, 19, 2019
|
|
5Y6T
 
 | | Crystal structure of endo-1,4-beta-mannanase from Eisenia fetida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, endo-1,4-beta-mannanase | | Authors: | Hirano, Y, Ueda, M, Tamada, T. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gene cloning, expression, and X-ray crystallographic analysis of a beta-mannanase from Eisenia fetida.
Enzyme.Microb.Technol., 117, 2018
|
|
2E0M
 
 | | Mutant Human Ribonuclease 1 (T24L, Q28L, R31L, R32L) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0O
 
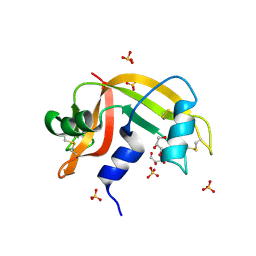 | | Mutant Human Ribonuclease 1 (V52L, D53L, N56L, F59L) | | Descriptor: | GLYCEROL, Ribonuclease, SULFATE ION | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0L
 
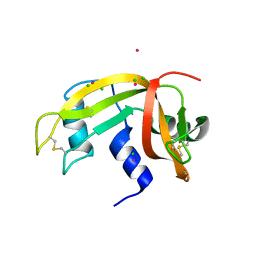 | | Mutant Human Ribonuclease 1 (Q28L, R31L, R32L) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0J
 
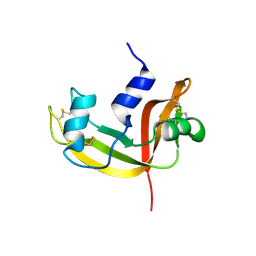 | | Mutant Human Ribonuclease 1 (R31L, R32L) | | Descriptor: | Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
