6J2Z
 
 | | AtFKBP53 N-terminal Nucleoplasmin Domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-03 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
3ZPE
 
 | |
3ZPF
 
 | |
3KRQ
 
 | | Crystal structure of the complex of lactoperoxidase with a potent inhibitor amino-triazole at 2.2a resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-AMINO-1,2,4-TRIAZOLE, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kushwaha, G.S, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-11-19 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | First structural evidence for the mode of diffusion of aromatic ligands and ligand-induced closure of the hydrophobic channel in heme peroxidases
J.Biol.Inorg.Chem., 15, 2010
|
|
3I6N
 
 | | Mode of Binding of the Tuberculosis Prodrug Isoniazid to Peroxidases: Crystal Structure of Bovine Lactoperoxidase with Isoniazid at 2.7 Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIAZENYLCARBONYL)PYRIDINE, CALCIUM ION, ... | | Authors: | Singh, A.K, Kumar, R.P, Pandey, N, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-07-07 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
2E9E
 
 | | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Prem kumar, R, Singh, N, Sharma, S, Singh, S.B, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution
To be Published
|
|
2DQK
 
 | | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Dey, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-05-29 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 resolution
To be Published
|
|
2DUJ
 
 | | Crystal structure of the complex formed between proteinase K and a synthetic peptide Leu-Leu-Phe-Asn-Asp at 1.67 A resolution | | Descriptor: | CALCIUM ION, LLFND, NITRATE ION, ... | | Authors: | Singh, A.K, Singh, N, Somvanshi, R.K, Gupta, D, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-23 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution
To be Published
|
|
2EFB
 
 | | Crystal structure of the complex of goat lactoperoxidase with phosphate at 2.94 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Hariprasad, G, Prem Kumar, R, Singh, N, Bhushan, A, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with phosphate at 2.94 A resolution
To be Published
|
|
2EHA
 
 | | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Ethayathulla, A.S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution
to be published
|
|
2DP4
 
 | | Crystal structure of the complex formed between proteinase K and a human lactoferrin fragment at 2.9 A resolution | | Descriptor: | 8-mer peptide from Lactotransferrin, Proteinase K | | Authors: | Singh, A.K, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-05-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex formed between proteinase K and a human lactoferrin fragment at 2.9 A resolution
To be Published
|
|
4D1G
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, second P212121 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4D1F
 
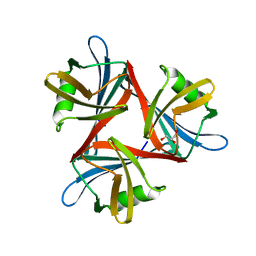 | |
4D0U
 
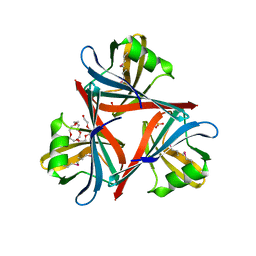 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, selenomethionine-derivative | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4D0V
 
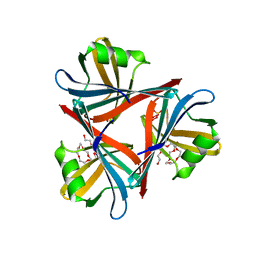 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, I213 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
7WIM
 
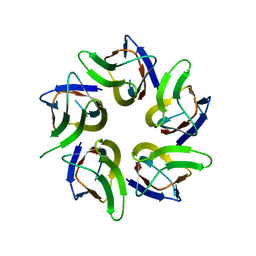 | |
7Z2H
 
 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2G
 
 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with doravirine | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, DNA (38-MER), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z24
 
 | | Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-mer), Reverse transcriptase/ribonuclease H | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2D
 
 | |
7Z29
 
 | | Cryo-EM structure of NNRTI resistant M184I/E138K mutant HIV-1 reverse transcriptase with a DNA aptamer in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (38-MER), Reverse transcriptase/ribonuclease H, ... | | Authors: | Singh, A.K, Das, K. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of wild-type and E138K/M184I mutant HIV-1 RT/DNA complexed with inhibitors doravirine and rilpivirine.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z2E
 
 | |
5NBH
 
 | |
5NC1
 
 | |
5N8D
 
 | |
