5XQL
 
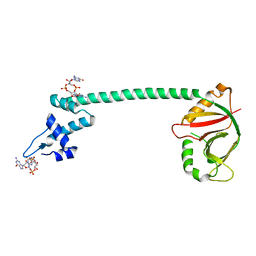 | | Crystal structure of a Pseudomonas aeruginosa transcriptional regulator | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Multidrug-efflux transporter 1 regulator | | Authors: | Raju, H, Sharma, R. | | Deposit date: | 2017-06-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Crystal structure of BrlR with c-di-GMP
Biochem. Biophys. Res. Commun., 490, 2017
|
|
2JWH
 
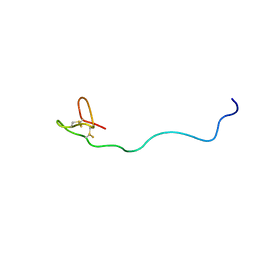 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2JWG
 
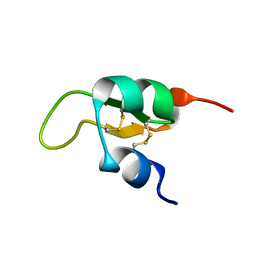 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
5YBC
 
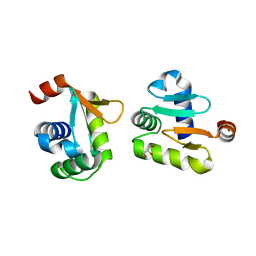 | | X-ray structure of native ETS-domain domain of Ergp55 | | Descriptor: | GLYCEROL, Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5YBD
 
 | | X-ray structure of ETS domain of Ergp55 in complex with E74DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7MBJ
 
 | |
6QIZ
 
 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
7PQE
 
 | |
5YC9
 
 | |
7JY3
 
 | | Structure of HbA with compound 23 (PF-07059013) | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-{(1S)-1-[(2-amino-6-fluoroquinolin-3-yl)oxy]ethyl}-5-(1H-pyrazol-1-yl)pyridin-2(1H)-one, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JXZ
 
 | | Structure of HbA with compound (S)-4 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JY1
 
 | | Structure of HbA with compound 19 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7JY0
 
 | | Structure of HbA with compound 9 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2-amino-3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}quinoline-6-carboxamide, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
4N7X
 
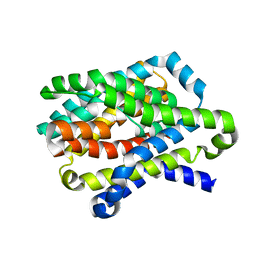 | |
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
7PPO
 
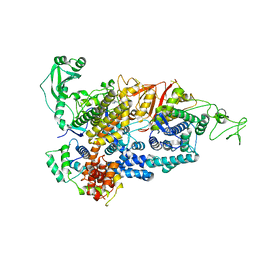 | |
7W22
 
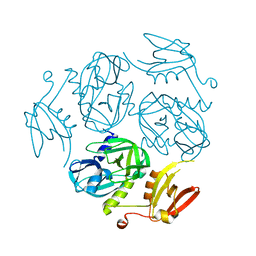 | | Structure of the M. tuberculosis HtrA K436A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W23
 
 | | Structure of the M. tuberculosis HtrA S363A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W24
 
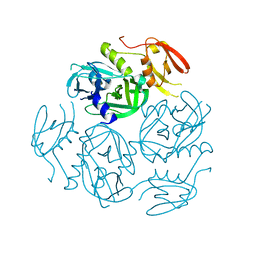 | | Structure of the M. tuberculosis HtrA N383A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W25
 
 | | Structure of the M. tuberculosis HtrA S413A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VZ0
 
 | | Structure of the M. tuberculosis HtrA S407A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W21
 
 | | Structure of the M. tuberculosis HtrA N269A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VYZ
 
 | | Structure of the M. tuberculosis HtrA S367A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W4R
 
 | |
7W4W
 
 | |
