2D38
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2YYS
 
 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
2ZZF
 
 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZZG
 
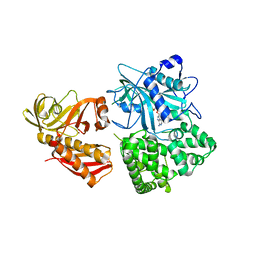 | | Crystal structure of alanyl-tRNA synthetase in complex with 5''-O-(N-(L-alanyl)-sulfamyoxyl) adenine without oligomerization domain | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2DCU
 
 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Translation initiation factor 2 beta subunit, ... | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2006-01-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2EK9
 
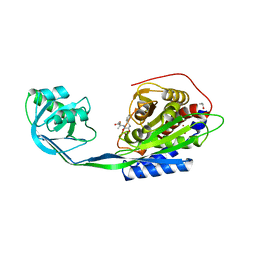 | |
2EK8
 
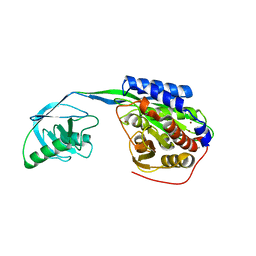 | |
2D74
 
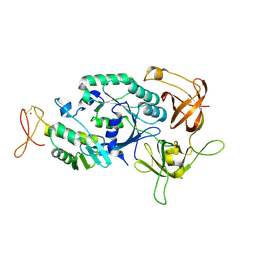 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer | | Descriptor: | Translation initiation factor 2 beta subunit, Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2005-11-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7EN4
 
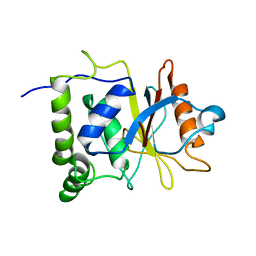 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
3WQ7
 
 | |
3WXP
 
 | |
3WT3
 
 | |
3WY6
 
 | |
3WQ0
 
 | |
3WQ1
 
 | |
3WR0
 
 | |
1V7O
 
 | |
3D7A
 
 | |
3WP3
 
 | |
3VGI
 
 | | The crystal structure of hyperthermophilic family 12 endocellulase from Pyrococcus furiosus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Endoglucanase A, ... | | Authors: | Kataoka, M, Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-08-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus.
Febs Lett., 586, 2012
|
|
3WQH
 
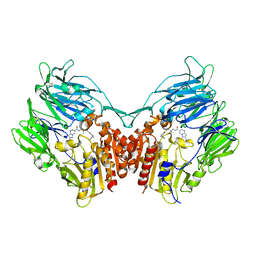 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | Authors: | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | Deposit date: | 2014-01-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
2AGT
 
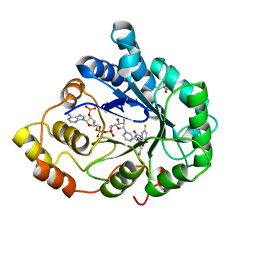 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
7F1I
 
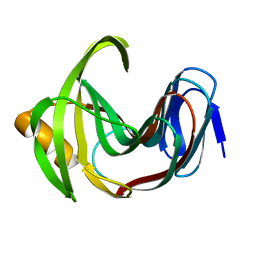 | | Designed enzyme RA61 M48K/I72D mutant: form II | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
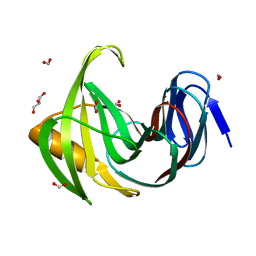 | | Designed enzyme RA61 M48K/I72D mutant: form I | | Descriptor: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
