7Z2K
 
 | | Crystal structure of SARS-CoV-2 Main Protease in orthorhombic space group p212121 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-02-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7Z3U
 
 | | Crystal structure of SARS-CoV-2 Main Protease after incubation with Sulfo-Calpeptin | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, Calpetin, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z3T
 
 | | Crystal structure of apo human Cathepsin L | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7Z58
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound Calpeptin | | Descriptor: | 1,2-ETHANEDIOL, Calpeptin, Cathepsin L, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7YAS
 
 | | HYDROXYNITRILE LYASE, LOW TEMPERATURE NATIVE STRUCTURE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (HYDROXYNITRILE LYASE), ... | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
1QJ4
 
 | |
4YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH CHLORALHYDRATE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION, TRI-CHLORO-ACETALDEHYDE | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
5YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH HEXAFLUOROACETONE | | Descriptor: | 1,1,1,3,3,3-HEXAFLUOROPROPANEDIOL, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
3YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH ACETONE | | Descriptor: | ACETONE, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
6YAS
 
 | | HYDROXYNITRILE LYASE FROM HEVEA BRASILIENSIS, ROOM TEMPERATURE STRUCTURE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
2YAS
 
 | |
487D
 
 | |
2VB0
 
 | | Crystal structure of coxsackievirus B3 proteinase 3C | | Descriptor: | CHLORIDE ION, POLYPROTEIN 3BCD | | Authors: | Anand, K, Mesters, J.R, Goerlach, R, Zell, R, Hilgenfeld, R. | | Deposit date: | 2007-09-05 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Coxsackie Virus B3 Proteinase 3C
To be Published
|
|
8Q4S
 
 | |
8QOB
 
 | |
2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SEX-LETHAL PROTEIN | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-07-16 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
3TPK
 
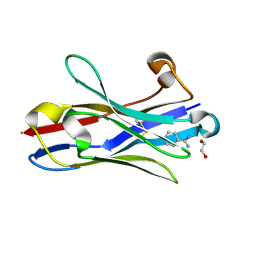 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
1C2X
 
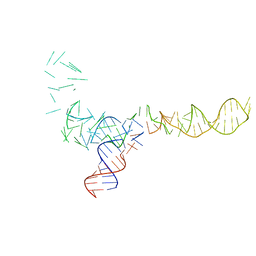 | |
1C2W
 
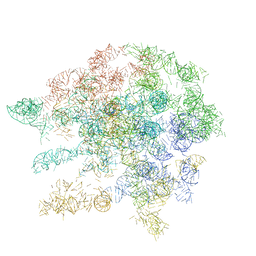 | |
1SXL
 
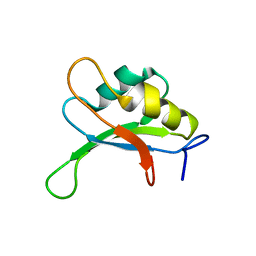 | |
1MNX
 
 | |
1SCI
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCQ
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
