4E50
 
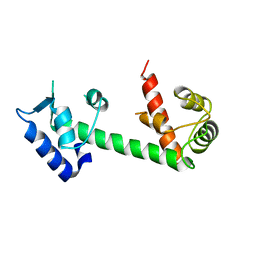 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
3T0I
 
 | |
3T7R
 
 | |
3SVZ
 
 | |
3T7S
 
 | |
3SXJ
 
 | |
3T7T
 
 | |
4KIR
 
 | |
4KQN
 
 | |
6K5P
 
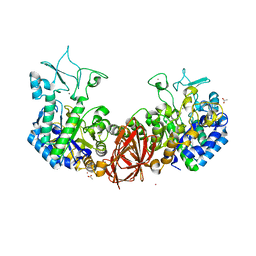 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
8GFF
 
 | |
8GFC
 
 | |
8GFH
 
 | |
8GFM
 
 | |
8GFQ
 
 | |
8GFE
 
 | |
8GFG
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with Z7912 inhibitor | | Descriptor: | (3S,3aR,5S,6S,6aS)-2-oxohexahydro-2H-3,5-methanocyclopenta[b]furan-6-yl 2-acetamido-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFS
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with siastatin B inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, CITRIC ACID, Lytic transglycosylase domain-containing protein | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GEZ
 
 | |
8GF0
 
 | |
8GFB
 
 | |
8GFL
 
 | |
8GFP
 
 | | Crystal structure of soluble lytic transglycosylase Cj0843 of Campylobacter jejuni in complex with N-acetyl-2,3-dehydro-2-deoxyneuraminic acid inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CITRIC ACID, Lytic transglycosylase domain-containing protein, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Exploring the inhibition of the soluble lytic transglycosylase Cj0843c of Campylobacter jejuni via targeting different sites with different scaffolds.
Protein Sci., 32, 2023
|
|
8GFD
 
 | |
8GFI
 
 | |
