6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M6Y
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M72
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5FHL
 
 | |
5FHJ
 
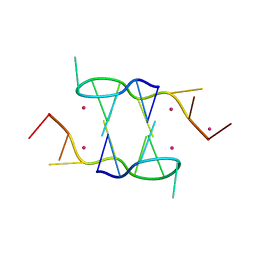 | |
4P77
 
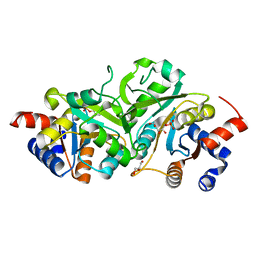 | | Structure of ribB complexed with substrate Ru5P | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, RIBULOSE-5-PHOSPHATE | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6D
 
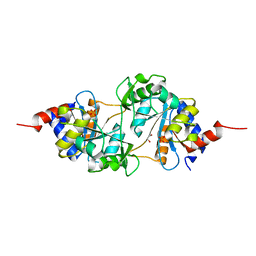 | | Structure of ribB complexed with PO4 ion | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, PHOSPHATE ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6P
 
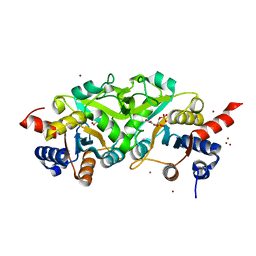 | | Structure of ribB complexed with inhibitor (4PEH) and metal ions | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ZINC ION | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P8J
 
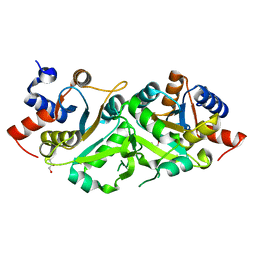 | | Structure of ribB | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
3MGZ
 
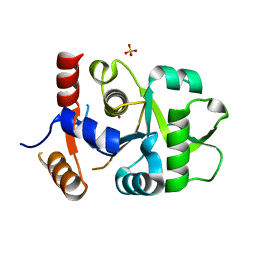 | | Crystal structure of DHBPS domain of bi-functional DHBPS/GTP cyclohydrolase II from Mycobacterium tuberculosis at pH 4.0 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION | | Authors: | Singh, M, Kumar, P, Karthikeyan, S. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MIO
 
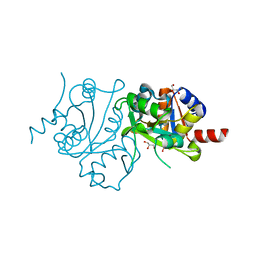 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis at pH 6.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
3MK5
 
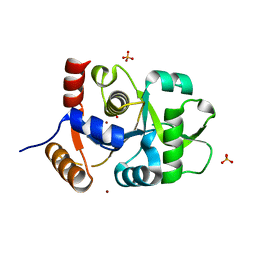 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis with sulfate and zinc at pH 4.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION, ZINC ION | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
5HWI
 
 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5I2U
 
 | | Crystal structure of a novel Halo-Tolerant Cellulase from Soil Metagenome | | Descriptor: | Cellulase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garg, R, Brahma, V, Srivastava, R, Verma, L, Karthikeyan, S, Sahni, G. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome
Sci Rep, 6, 2016
|
|
4RL4
 
 | |
4I14
 
 | | Crystal Structure of Mtb-ribA2 (Rv1415) | | Descriptor: | Riboflavin biosynthesis protein RibBA, SULFATE ION, ZINC ION | | Authors: | Singh, M, Kumar, P, Yadav, S, Gautam, R, Sharma, N, Karthikeyan, S. | | Deposit date: | 2012-11-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure reveals the molecular mechanism of bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II (Rv1415) from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8H5S
 
 | | Crystal structure of Rv3400 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, CHLORIDE ION, ... | | Authors: | Singh, L, Karthikeyan, S, Thakur, K.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization reveals Rv3400 codes for beta-phosphoglucomutase in Mycobacterium tuberculosis.
Protein Sci., 33, 2024
|
|
4OW8
 
 | | Crystal structure of kinase domain of PknA from Mtb | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PknA | | Authors: | Ravala, S.K, Singh, S, Yadav, G.S, Karthikeyan, S, Chakraborti, P.K. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evidence that phosphorylation of threonine in the GT motif triggers activation of PknA, a eukaryotic-type serine/threonine kinase from Mycobacterium tuberculosis.
Febs J., 282, 2015
|
|
1DQ7
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NEUROTOXIN FROM RED SCORPION (BUTHUS TAMULUS) AT 2.2A RESOLUTION. | | Descriptor: | NEUROTOXIN | | Authors: | Sharma, M, Yadav, S, Karthikeyan, S, Kumar, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-30 | | Release date: | 2000-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional Structure of a Neurotoxin from Red Scorpion (Buthus tamulus) at 2.2A Resolution
To be Published
|
|
1BJR
 
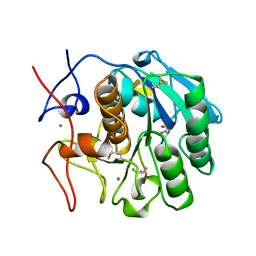 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
8BR4
 
 | |
4P8E
 
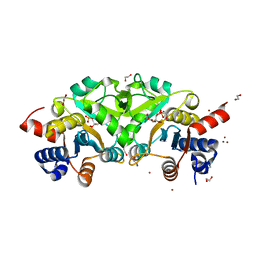 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
4P6C
 
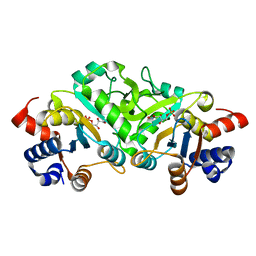 | | Structure of ribB complexed with inhibitor 4PEH | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
1CNM
 
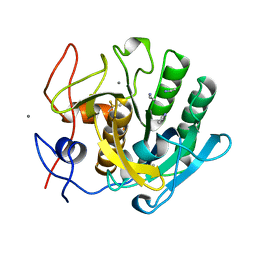 | | ENHANCEMENT OF CATALYTIC EFFICIENCY OF PROTEINASE K THROUGH EXPOSURE TO ANHYDROUS ORGANIC SOLVENT AT 70 DEGREES CELSIUS | | Descriptor: | ACETONITRILE, CALCIUM ION, PROTEIN (PROTEINASE K) | | Authors: | Gupta, M.N, Tyagi, R, Sharma, S, Karthikeyan, S, Singh, T.P. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enhancement of catalytic efficiency of enzymes through exposure to anhydrous organic solvent at 70 degrees C. Three-dimensional structure of a treated serine proteinase at 2.2 A resolution.
Proteins, 39, 2000
|
|
3VOS
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With glycerol and sulfate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
