4QZV
 
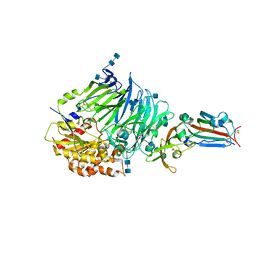 | | Bat-derived coronavirus HKU4 uses MERS-CoV receptor human CD26 for cell entry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Gao, F.G, Wang, Q.H, Qi, J.X, Lu, G.W. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Bat Origins of MERS-CoV Supported by Bat Coronavirus HKU4 Usage of Human Receptor CD26.
Cell Host Microbe, 16, 2014
|
|
6LW2
 
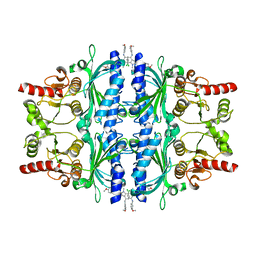 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
8XQF
 
 | | Cryo-EM structure of human monomeric APJR-Gi complex with apelin-13. | | Descriptor: | Apelin-13, G protein subunit alpha i1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-01-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8XQE
 
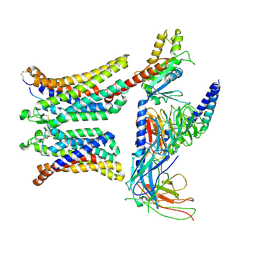 | | Cryo-EM structure of human dimeric APJR-Gi complex with apelin-13. | | Descriptor: | Apelin-13, G protein subunit alpha i1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-01-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8XQI
 
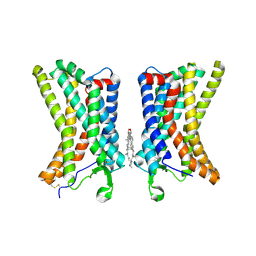 | | Cryo-EM structure of human dimeric Apelin receptor. | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,Apelin receptor | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-01-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8XQJ
 
 | | Cryo-EM structure of human dimeric APJR complex with antagonistic antibody | | Descriptor: | CHOLESTEROL, JN241, Soluble cytochrome b562,Apelin receptor | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-01-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8Z7J
 
 | | Cryo-EM structure of APJR-Gi complex with agonistic antibody | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8Z74
 
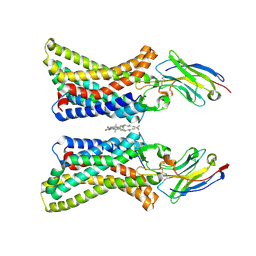 | | Cryo-EM structure of APJR complex with agonistic antibody | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,Apelin receptor, agonistic antibody | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F. | | Deposit date: | 2024-04-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the regulation of monomeric and dimeric apelin receptor.
Nat Commun, 16, 2025
|
|
8YPD
 
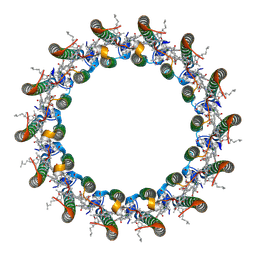 | | Cryo-EM structure of the LH1 complex from Allochromatium tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Beta subunit of light-harvesting 1 complex, LH1 alpha subunit, ... | | Authors: | Wang, G.-L, Sun, S, Yu, L.-J. | | Deposit date: | 2024-03-16 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Probing the Dual Role of Ca 2+ in the Allochromatium tepidum LH1-RC Complex by Constructing and Analyzing Ca 2+ -Bound and Ca 2+ -Free LH1 Complexes.
Biomolecules, 15, 2025
|
|
8YPB
 
 | | Cryo-EM structure of the LH1 complex from Allochromatium tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Beta subunit of light-harvesting 1 complex, CALCIUM ION, ... | | Authors: | Wang, G.-L, Sun, S, Yu, L.-J. | | Deposit date: | 2024-03-16 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Probing the Dual Role of Ca 2+ in the Allochromatium tepidum LH1-RC Complex by Constructing and Analyzing Ca 2+ -Bound and Ca 2+ -Free LH1 Complexes.
Biomolecules, 15, 2025
|
|
8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
8ZE6
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to MjDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-04 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
7EJU
 
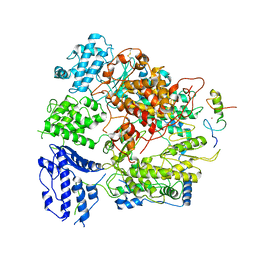 | | Junin virus(JUNV) RNA polymerase L complexed with Z protein | | Descriptor: | MAGNESIUM ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Chen, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2021-07-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for recognition and regulation of arenavirus polymerase L by Z protein.
Nat Commun, 12, 2021
|
|
7EZP
 
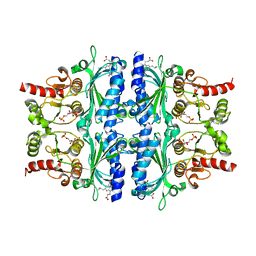 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
