3A23
 
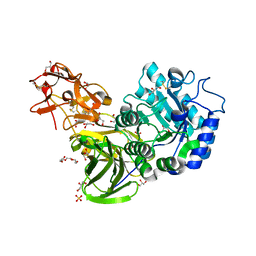 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with D-galactose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
3A21
 
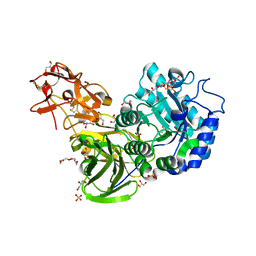 | | Crystal Structure of Streptomyces avermitilis beta-L-Arabinopyranosidase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
3WFL
 
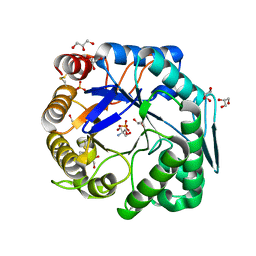 | | Crtstal structure of glycoside hydrolase family 5 beta-mannanase from Talaromyces trachyspermus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Suzuki, K, Ichinose, H, Kamino, K, Ogasawara, W, Kaneko, S, Fushinobu, S. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, cloning, functional expression, structure, and characterization of a thermostable beta-mannanase from Talaromyces trachyspermus and its efficiency in production of mannooligosaccharides from coffee wastes
To be Published
|
|
3AB8
 
 | |
3AB7
 
 | |
1IZE
 
 | | Crystal structure of Aspergillus oryzae Aspartic proteinase complexed with pepstatin | | Descriptor: | Pepstatin, alpha-D-mannopyranose, aspartic proteinase | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Aspergillus oryzae aspartic proteinase and its complex with an inhibitor pepstatin at 1.9A resolution.
J.Mol.Biol., 326, 2003
|
|
1IZD
 
 | | Crystal structure of Aspergillus oryzae Aspartic Proteinase | | Descriptor: | Aspartic proteinase, alpha-D-mannopyranose | | Authors: | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Aspergillus oryzae Aspartic Proteinase and its Complex with an Inhibitor Pepstatin at 1.9 A Resolution
J.Mol.Biol., 326, 2003
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
8HMN
 
 | |
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
6KRN
 
 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, Mating factor alpha,GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6KRL
 
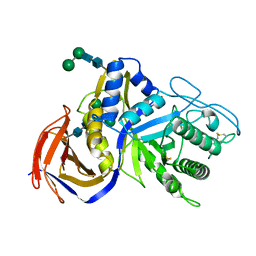 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6M5Z
 
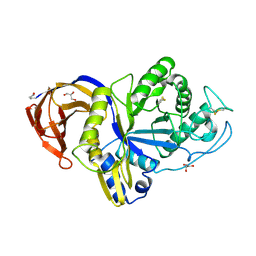 | | Catalytic domain of GH30 xylanase C from Talaromyces cellulolyticus | | Descriptor: | ACETATE ION, GH30 Xylanase C, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2020-03-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7VF8
 
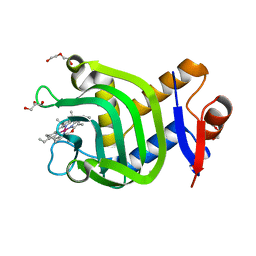 | | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation | | Descriptor: | CITRIC ACID, Co-5-octaethyloxaporphyrinium cation, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation
To Be Published
|
|
7VF7
 
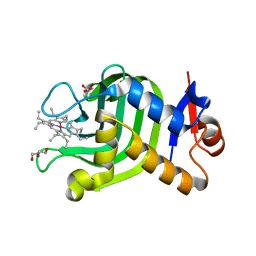 | | Crystal Structure of HasAp with Co-octaethylporphyrin | | Descriptor: | Co-octaethylporphyrin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-octaethylporphyrin
To Be Published
|
|
6IQM
 
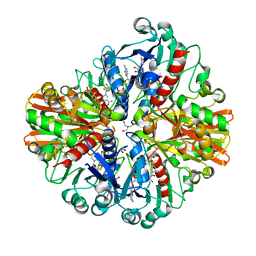 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with NAD+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
6IQV
 
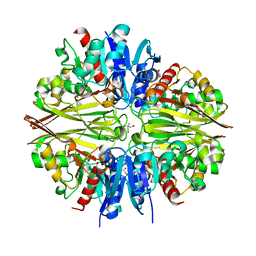 | | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase Complexed with Hg2+ from Lactobacillus plantarum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Yoneda, K, Kinoshita, H. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Cell Surface Glyceraldehyde-3-Phosphate Dehydrogenase from Lactobacillus plantarum: Insight into the Mercury Binding Mechanism
Milk Sci, 68, 2019
|
|
6IUJ
 
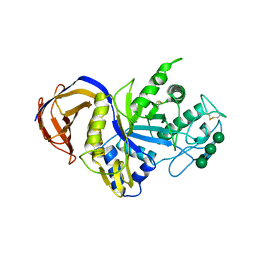 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2018-11-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of a bifunctional GH30-7 xylanase B from the filamentous fungusTalaromyces cellulolyticus.
J. Biol. Chem., 294, 2019
|
|
2RQ2
 
 | | The solution structure of the N-terminal fragment of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Mizuguchi, M, Aizawa, T, Shinoda, H, Demura, M, Kawabata, S, Kawano, K. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: big defensin changes its N-terminal structure to associate with the target membrane
Biochemistry, 48, 2009
|
|
5B6F
 
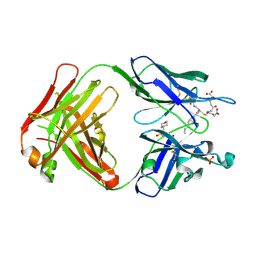 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
5X7S
 
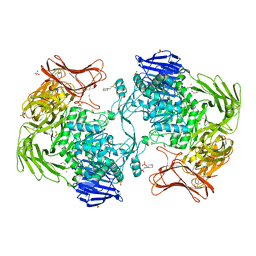 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7Q
 
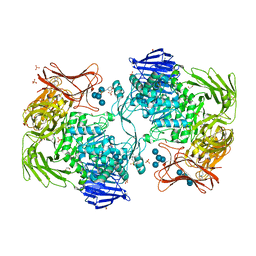 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7O
 
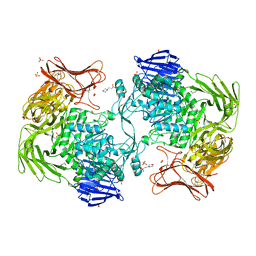 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Suzuki, N, Kishine, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
