4K1X
 
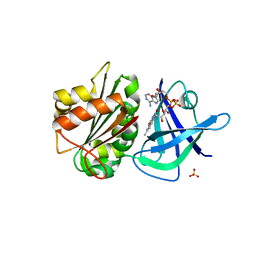 | | Ferredoxin-NADP(H) Reductase mutant with Ala 266 replaced by Tyr (A266Y) and residues 267-272 deleted. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:ferredoxin reductase, SULFATE ION | | Authors: | Bortolotti, A, Sanchez-Azqueta, A, Maya, C.M, Velazquez-Campoy, A, Hermoso, J.A, Cortez, N. | | Deposit date: | 2013-04-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal extension of bacterial flavodoxin-reductases: Involvement in the hydride transfer mechanism from the coenzyme.
Biochim.Biophys.Acta, 1837, 2013
|
|
4UPD
 
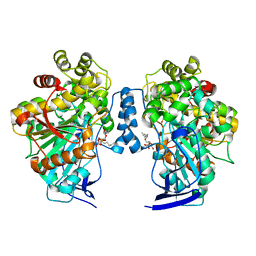 | | Open conformation of O. piceae sterol esterase mutant I544W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, STEROL ESTERASE, TRIETHYLENE GLYCOL, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
5A8A
 
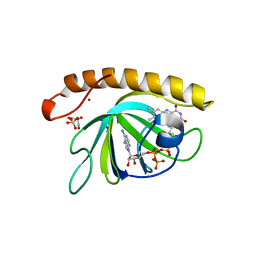 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP (P3 2 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AA3
 
 | |
5A5X
 
 | |
4BE9
 
 | | Open conformation of O. piceae sterol esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
4BEQ
 
 | |
4BJ4
 
 | | Structure of Pseudomonas aeruginosa amidase Ampdh2 | | Descriptor: | AMPDH2, CITRATE ANION | | Authors: | Martinez-Caballero, C.S, Carrasco-Lopez, C, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4BF5
 
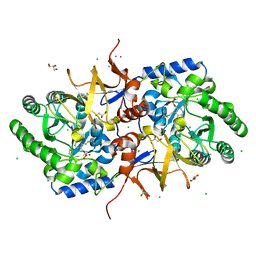 | |
4BL2
 
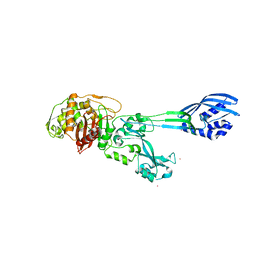 | |
4BHY
 
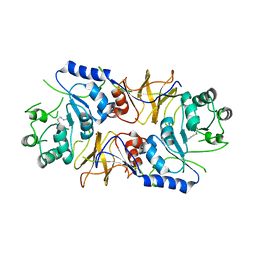 | | Structure of alanine racemase from Aeromonas hydrophila | | Descriptor: | ALANINE RACEMASE | | Authors: | Otero, L.H, Carrasco-Lopez, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-04-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2J8F
 
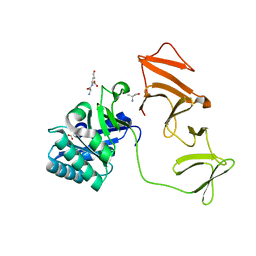 | |
2IXU
 
 | |
2IXV
 
 | |
2J8G
 
 | |
8A39
 
 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
2YP6
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
2Y8P
 
 | | Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli | | Descriptor: | ENDO-TYPE MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE A | | Authors: | Artola-Recolons, C, Carrasco-Lopez, C, Llarrull, L.I, Kumarasiri, M, Lastochkin, E, Martinez-Ilarduya, I, Meindl, K, Uson, I, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-Resolution Crystal Structure of Mlte, an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase from Escherichia Coli.
Biochemistry, 50, 2011
|
|
8B58
 
 | |
3ESX
 
 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
2BGI
 
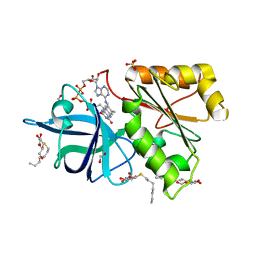 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus complexed with three molecules of the detergent n-heptyl- beta-D-thioglucoside at 1.7 Angstroms | | Descriptor: | CARBON DIOXIDE, FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Ferredoxin-Nadp(H) Reductase from Rhodobacter Capsulatus: Molecular Structure and Catalytic Mechanism
Biochemistry, 44, 2005
|
|
2BGJ
 
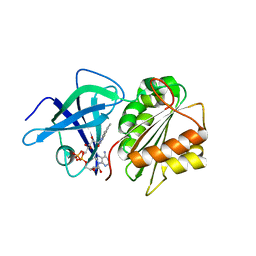 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus at 2.1 Angstroms | | Descriptor: | FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ferredoxin-NADP(H) reductase from Rhodobacter capsulatus: molecular structure and catalytic mechanism.
Biochemistry, 44, 2005
|
|
4MPI
 
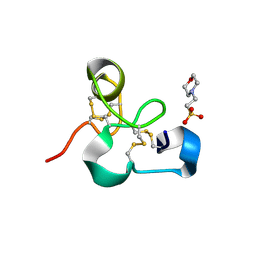 | | Crystal structure of the chitin-binding module (CBM18) of a chitinase-like protein from Hevea brasiliensis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Class I chitinase | | Authors: | Martinez-Caballero, C.S, Hermoso, J.A, Rodriguez-Romero, A. | | Deposit date: | 2013-09-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Comparative study of two GH19 chitinase-like proteins from Hevea brasiliensis, one exhibiting a novel carbohydrate-binding domain.
Febs J., 281, 2014
|
|
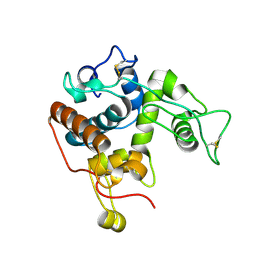
4MST
 
 | |
