1UUB
 
 | |
4XAH
 
 | |
3E6U
 
 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3E73
 
 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
4YDU
 
 | | Crystal structure of E. coli YgjD-YeaZ heterodimer in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W, Collinet, B, Perrochia, L, Durand, D, Van Tilbeurgh, H. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW7
 
 | |
4WX8
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WXA
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1, ... | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WQ4
 
 | | E. coli YgjD(E12A)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, W, Collinet, B. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4WW5
 
 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW9
 
 | |
4X3X
 
 | | The crystal structure of Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
1UUA
 
 | |
5DHB
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5DHC
 
 | | Cooperativity and Downstream Binding in RNA Replication | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Fahrenbach, A.C, Tam, C.P, Szostak, J.W. | | Deposit date: | 2015-08-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5TGP
 
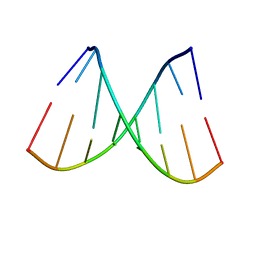 | | DNA 8mer containing two 2SeT modifications | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*(2ST))-R(P*G)-D(P*(2ST))-R(P*AP*CP*AP*C)-3') | | Authors: | Zhang, W, Huang, Z. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA 8mer containing two 2SeT modifications
To Be Published
|
|
5L00
 
 | | Self-complimentary RNA 15mer binding with GMP monomers | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*G)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
8SPH
 
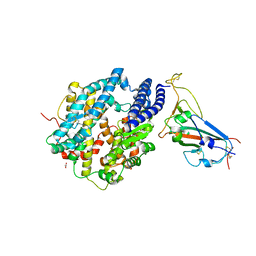 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
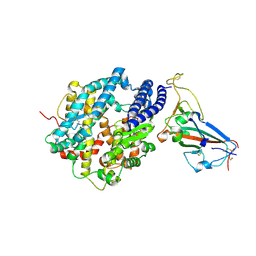 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
5WCO
 
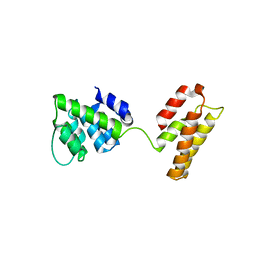 | | Matrix Protein (M1) of Infectious Salmon Anaemia Virus | | Descriptor: | NS2 | | Authors: | Zhang, W, Zheng, W, Toh, Y, Betancourt, M.A, Tu, J, Fan, Y, Vakharia, V, Liu, J, McNew, J.A, Jin, M, Tao, Y.J. | | Deposit date: | 2017-07-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of an orthomyxovirus matrix protein reveals mechanisms for self-polymerization and membrane association.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KRG
 
 | | RNA 15mer duplex binding with PZG monomer | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*G)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
