3OBU
 
 | |
3OBQ
 
 | |
3OBX
 
 | |
3OBS
 
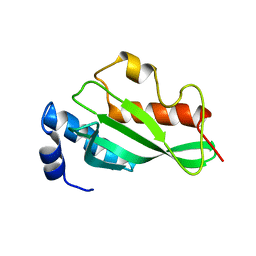 | | Crystal structure of Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Im, Y.J, Hurley, J.H. | | Deposit date: | 2010-08-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic and Functional Analysis of the ESCRT-I /HIV-1 Gag PTAP Interaction.
Structure, 18, 2010
|
|
5OSI
 
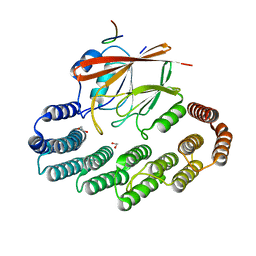 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSH
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OT4
 
 | | Structure of the Legionella pneumophila effector RidL (1-866) | | Descriptor: | GLYCEROL, Interaptin | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2FIF
 
 | |
2FID
 
 | |
