5FQ8
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-decanoyloxypropyl decanoate, BT_2262 (UNCHARACTERISED LIPOPROTEIN), ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ7
 
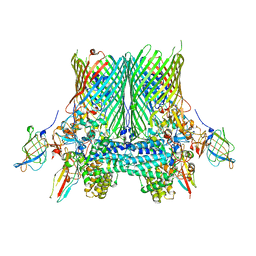 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2261, BT_2262, BT_2263, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ6
 
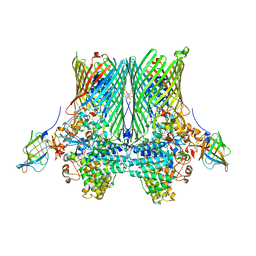 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | 3-decanoyloxypropyl decanoate, BT_2261, CALCIUM ION, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ4
 
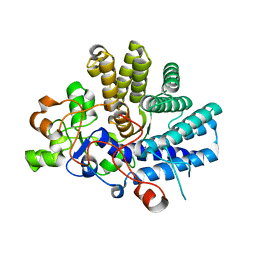 | | Crystal structure of the lipoprotein BT2263 from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, PUTATIVE LIPOPROTEIN | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5FQ3
 
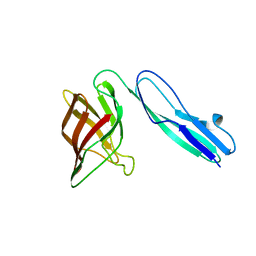 | | Crystal structure of the lipoprotein BT2262 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2262 | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
5G2U
 
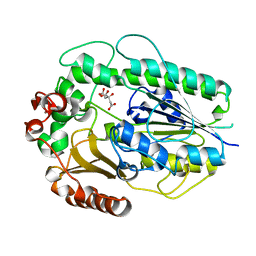 | | Structure of BT1596,a 2-O GAG sulfatase | | Descriptor: | 2-O GLYCOSAMINOGLYCAN SULFATASE, CITRIC ACID, ZINC ION | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2T
 
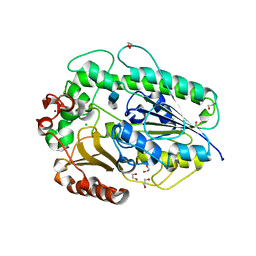 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2V
 
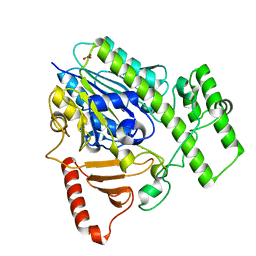 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2X7X
 
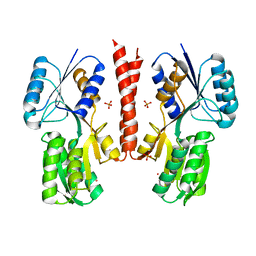 | | Fructose binding periplasmic domain of hybrid two component system BT1754 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SENSOR PROTEIN, ... | | Authors: | Sonnenburg, E.D, Zheng, H, Joglekar, P, Higginbottom, S, Firbank, S.J, Bolam, D.N, Sonnenburg, J.L. | | Deposit date: | 2010-03-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations.
Cell(Cambridge,Mass.), 141, 2010
|
|
2XFD
 
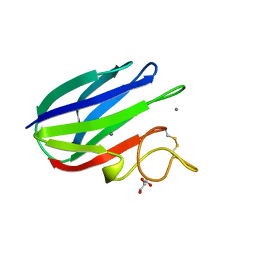 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2W9X
 
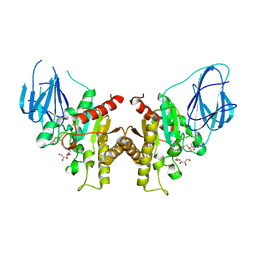 | | The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions | | Descriptor: | GLYCEROL, PUTATIVE ACETYL XYLAN ESTERASE | | Authors: | Montanier, C, Money, V.A, Pires, V, Flint, J.E, Benedita, P.A, Goyal, A, Prates, J.A, Izumi, A, Stalbrand, H, Morland, C, Cartmell, A, Kolenova, K, Topakas, E, Dobson, E, Bolam, D.N, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2XBD
 
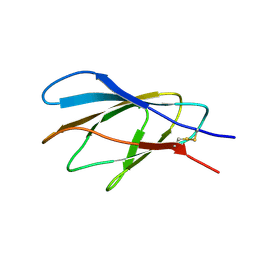 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-27 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2WAO
 
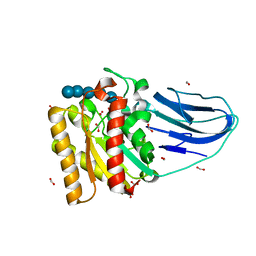 | | Structure of a family two carbohydrate esterase from Clostridium thermocellum in complex with cellohexaose | | Descriptor: | ENDOGLUCANASE E, FORMIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-10 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2WAB
 
 | | Structure of an active site mutant of a family two carbohydrate esterase from Clostridium thermocellum in complex with celluohexase | | Descriptor: | ENDOGLUCANASE E, GLYCEROL, IODIDE ION, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2WAA
 
 | | Structure of a family two carbohydrate esterase from Cellvibrio japonicus | | Descriptor: | ACETATE ION, GLYCEROL, XYLAN ESTERASE, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1GNY
 
 | | xylan-binding module CBM15 | | Descriptor: | SODIUM ION, XYLANASE 10C, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Szabo, S, Jamal, S, Xie, H, Charnock, S.J, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of a Family 15 Carbohydrate-Binding Module in Complex with Xylopentaose: Evidence that Xylan Binds in an Approximate Three-Fold Helical Conformation
J.Biol.Chem., 276, 2001
|
|
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8U
 
 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8T
 
 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
