6U05
 
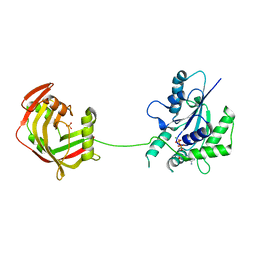 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
3M4A
 
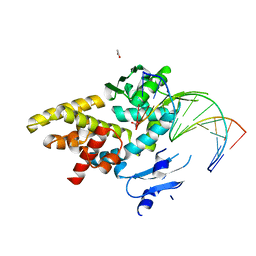 | | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site | | Descriptor: | ACETIC ACID, DNA (5'-D(*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*CP*CP*TP*TP*AP*TP*TP*C)-3'), ... | | Authors: | Patel, A, Yakovleva, L, Shuman, S, Mondragon, A. | | Deposit date: | 2010-03-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site.
Structure, 18, 2010
|
|
1S68
 
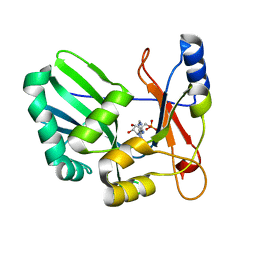 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
7SJR
 
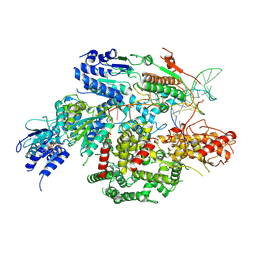 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
9D8A
 
 | |
3BGY
 
 | |
4W7S
 
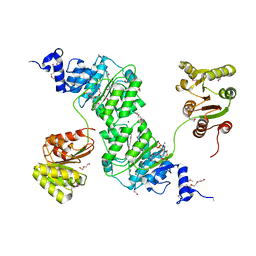 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
4WAN
 
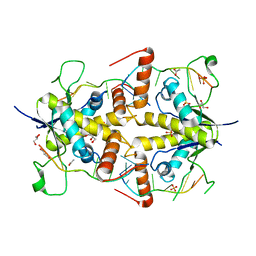 | | Crystal structure of Msl5 protein in complex with RNA at 1.8 A | | Descriptor: | ACETATE ION, Branchpoint-bridging protein, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4WAL
 
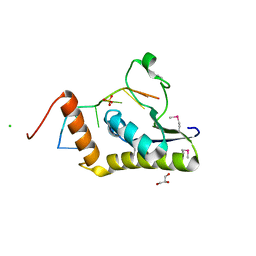 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
1VS0
 
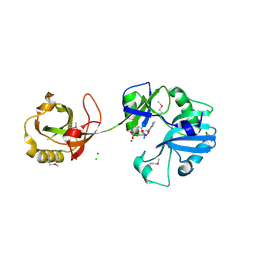 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
1Z3C
 
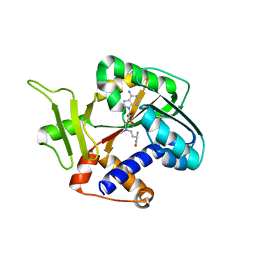 | | Encephalitozooan cuniculi mRNA Cap (Guanine-N7) Methyltransferasein complexed with AzoAdoMet | | Descriptor: | S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, mRNA CAPPING ENZYME | | Authors: | Hausmann, S, Zhang, S, Fabrega, C, Schneller, S.W, Lima, C.D, Shuman, S. | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis.
J.Biol.Chem., 280, 2005
|
|
9DO4
 
 | |
1YN9
 
 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
4CKB
 
 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4CKE
 
 | | Vaccinia virus capping enzyme complexed with SAH in P1 form | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4CKC
 
 | | Vaccinia virus capping enzyme complexed with SAH (monoclinic form) | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
1D8H
 
 | | X-RAY CRYSTAL STRUCTURE OF YEAST RNA TRIPHOSPHATASE IN COMPLEX WITH SULFATE AND MANGANESE IONS. | | Descriptor: | MANGANESE (II) ION, SULFATE ION, mRNA TRIPHOSPHATASE CET1 | | Authors: | Lima, C.D, Wang, L.K, Shuman, S. | | Deposit date: | 1999-10-24 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus.
Cell(Cambridge,Mass.), 99, 1999
|
|
6N0V
 
 | | tRNA ligase | | Descriptor: | MANGANESE (II) ION, tRNA ligase | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
6N0T
 
 | | tRNA ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
7T7N
 
 | | Structure of SPCC1393.13 protein from fission yeast | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Damage-control phosphatase SPCC1393.13, PHOSPHATE ION | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
9DRZ
 
 | |
9DQS
 
 | |
7T7O
 
 | |
7T7K
 
 | | Structure of SPAC806.04c protein from fission yeast bound to Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Damage-control phosphatase SPAC806.04c, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
6E33
 
 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
