1L4V
 
 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1MKC
 
 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1Q1O
 
 | | Solution Structure of the PB1 Domain of Cdc24p (Long Form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Kohjima, M, Ogura, K, Yokochi, M, Takeya, R, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-07-22 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PB1 domain and the PC motif-containing region are structurally similar protein binding modules
EMBO J., 22, 2003
|
|
2YZ0
 
 | |
2HSP
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
2Z0D
 
 | |
2Z0E
 
 | |
2ZPN
 
 | |
3A7P
 
 | |
3A77
 
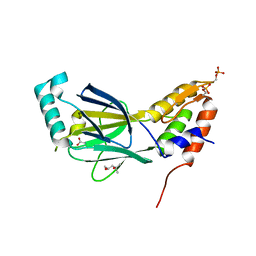 | | The crystal structure of phosphorylated IRF-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Interferon regulatory factor 3 | | Authors: | Takahasi, K, Horiuchi, M, Noda, N.N, Inagaki, F. | | Deposit date: | 2009-09-17 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change.
Genes Cells, 15, 2010
|
|
2ZZP
 
 | |
3A7O
 
 | |
1TCG
 
 | |
1TCK
 
 | |
1TCJ
 
 | |
1TCH
 
 | |
1WMH
 
 | | Crystal structure of a PB1 domain complex of Protein kinase c iota and Par6 alpha | | Descriptor: | Partitioning defective-6 homolog alpha, Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Suzuki, N.N, Horiuchi, M, Kohjima, M, Takeya, R, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Cell Polarity Regulator, a Complex between Atypical PKC and Par6 PB1 Domains
J.Biol.Chem., 280, 2005
|
|
1WZ3
 
 | | The crystal structure of plant ATG12 | | Descriptor: | autophagy 12b | | Authors: | Suzuki, N.N, Yoshimoto, K, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of plant ATG12 and its biological implication in autophagy.
Autophagy, 1, 2005
|
|
1X0N
 
 | | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor | | Descriptor: | 4-[(10S,14S,18S)-18-(2-AMINO-2-OXOETHYL)-14-(1-NAPHTHYLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL]BENZYLPHOSPHONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Ogura, K, Shiga, T, Yuzawa, S, Yokochi, M, Burke, T.R, Inagaki, F. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor
To be Published
|
|
1WLZ
 
 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | Descriptor: | CAP-binding protein complex interacting protein 1 isoform a | | Authors: | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | Deposit date: | 2004-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
1AHM
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
