3UXJ
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ0 | | Descriptor: | 1,2-ETHANEDIOL, 7-DEAZA-7-AMINOMETHYL-GUANINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Zhang, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from
Vibrio cholerae complexed with NADP and PreQ0
To be Published, 2012
|
|
6IF2
 
 | | Complex structure of Rab35 and its effector RUSC2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Iporin, MAGNESIUM ION, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6J4C
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 10 mM ZnSO4 | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
6JDT
 
 | |
6JE2
 
 | |
6JE0
 
 | |
6JDZ
 
 | |
6JE1
 
 | |
6IF3
 
 | | Complex structure of Rab35 and its effector ACAP2 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lin, L, Zhu, J, Zhang, R. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rab35/ACAP2 and Rab35/RUSC2 Complex Structures Reveal Molecular Basis for Effector Recognition by Rab35 GTPase.
Structure, 27, 2019
|
|
6J4D
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under pH 4.7, without Zn | | Descriptor: | CITRATE ANION, Cupin superfamily protein, GLYCEROL | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for the Isomerization Mechanism of MarH
To Be Published
|
|
6J4B
 
 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 400 mM Zinc acetate | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
6JEC
 
 | |
6JKL
 
 | |
6JDY
 
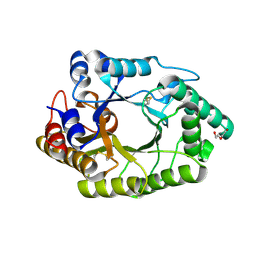 | |
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTK
 
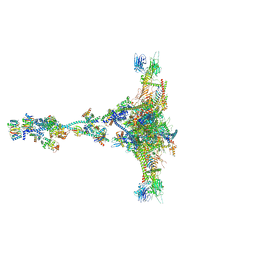 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6R4D
 
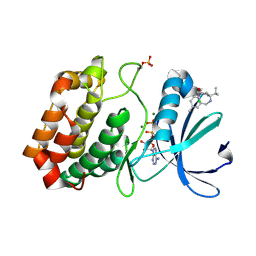 | | Aurora-A in complex with shape-diverse fragment 58 | | Descriptor: | (1~{S},10~{S})-12-cyclobutyl-5-methyl-1-oxidanyl-10-propan-2-yl-9,12-diazatricyclo[8.2.1.0^{2,7}]trideca-2(7),3,5-trien-11-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
6R4B
 
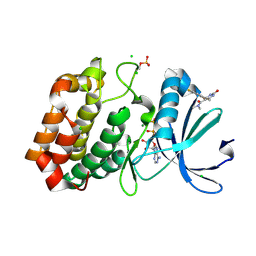 | | Aurora-A in complex with shape-diverse fragment 56 | | Descriptor: | (6~{S})-6-[2,4-bis(fluoranyl)phenyl]-~{N},~{N},4-trimethyl-2-oxidanylidene-5,6-dihydro-1~{H}-pyrimidine-5-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
6R49
 
 | | Aurora-A in complex with shape-diverse fragment 39 | | Descriptor: | (1~{S},10~{S})-12-cyclopropyl-1-oxidanyl-10-propan-2-yl-9,12-diazatricyclo[8.2.1.0^{2,7}]trideca-2(7),3,5-trien-11-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
5DD6
 
 | |
5DD3
 
 | |
5DD5
 
 | |
6R4C
 
 | | Aurora-A in complex with shape-diverse fragment 57 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
6R4A
 
 | | Aurora-A in complex with shape-diverse fragment 55 | | Descriptor: | 2-(benzimidazol-1-yl)-~{N}-(2-phenylethyl)ethanamide, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
8I7X
 
 | | Crystal structure of human ClpP in complex with ZG36 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists.
Eur.J.Med.Chem., 258, 2023
|
|
