5ZVV
 
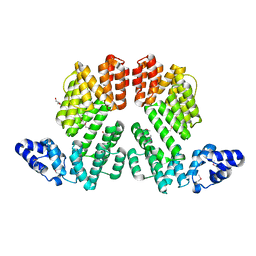 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
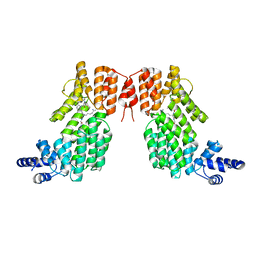 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
4HAX
 
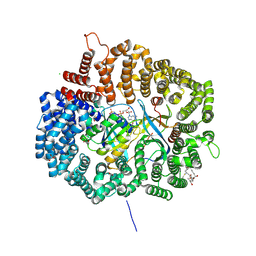 | |
4HB0
 
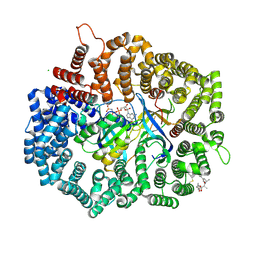 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAV
 
 | |
4HAU
 
 | |
4HB2
 
 | | Crystal structure of CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZW5
 
 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
6OCG
 
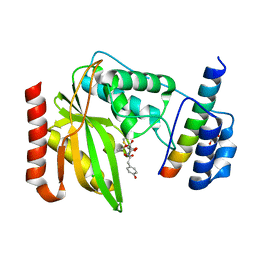 | | Crystal structure of VASH1-SVBP complex bound with EpoY | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4EBY
 
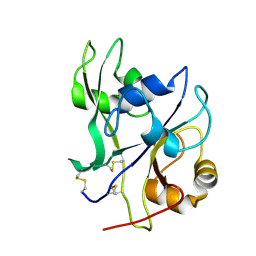 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBZ
 
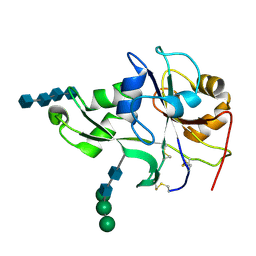 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
5ZVW
 
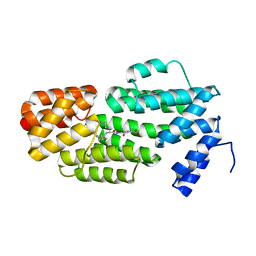 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZXL
 
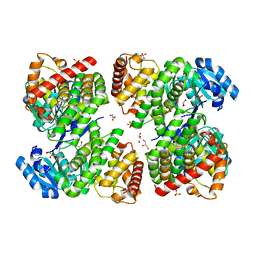 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
7WL2
 
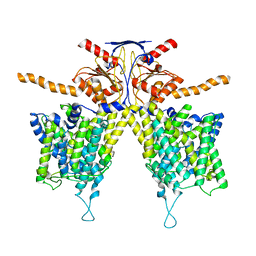 | |
7WLE
 
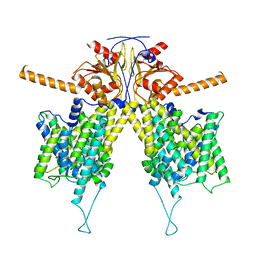 | |
7WL9
 
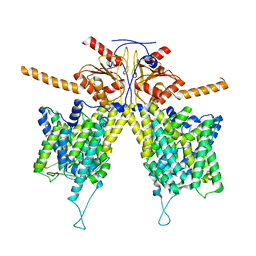 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WLA
 
 | |
7WK7
 
 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WK1
 
 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WL7
 
 | |
7WL8
 
 | |
7WLB
 
 | |
4HAW
 
 | |
4HB4
 
 | |
4JE0
 
 | |
