4J1R
 
 | | Crystal Structure of GSK3b in complex with inhibitor 15R | | Descriptor: | (2R)-2-(1H-indol-3-ylmethyl)-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
1QCO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
4J71
 
 | | Crystal Structure of GSK3b in complex with inhibitor 1R | | Descriptor: | (2R)-2-methyl-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, CHLORIDE ION, Glycogen synthase kinase-3 beta, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
1QQJ
 
 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
4P66
 
 | | Electrostatics of Active Site Microenvironments of E. coli DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
1QCN
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
4P68
 
 | | Electrostatics of Active Site Microenvironments for E. coli DHFR | | Descriptor: | ACETATE ION, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
1S1G
 
 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
6K9K
 
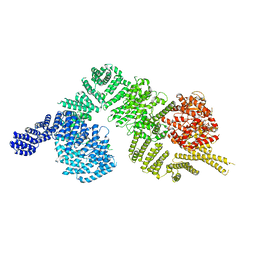 | | Monomeric human ATM (Ataxia telangiectasia mutated) kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Yuriy, C, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
2BI0
 
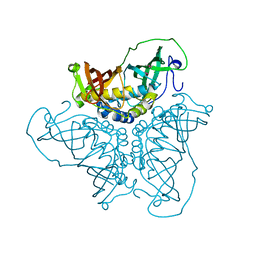 | | RV0216, A conserved hypothetical protein from Mycobacterium tuberculosis that is essential for bacterial survival during infection, has a double hotdogfold | | Descriptor: | CHLORIDE ION, HYPOTHETICAL PROTEIN RV0216 | | Authors: | Castell, A, Johansson, P, Unge, T, Jones, T.A, Backbro, K. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rv0216, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis that is Essential for Bacterial Survival During Infection, Has a Double Hotdog Fold
Protein Sci., 14, 2005
|
|
6K9L
 
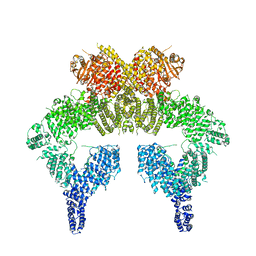 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
2K0Q
 
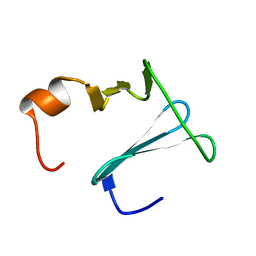 | | Solution structure of CopK, a periplasmic protein involved in copper resistance in Cupriavidus metallidurans CH34 | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Bersch, B, Favier, A, Schanda, P, Coves, J, van Aelst, S, Vallaeys, T, Wattiez, R, Mergeay, M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular structure and metal-binding properties of the periplasmic CopK protein expressed in Cupriavidus metallidurans CH34 during copper challenge.
J.Mol.Biol., 380, 2008
|
|
2L3Z
 
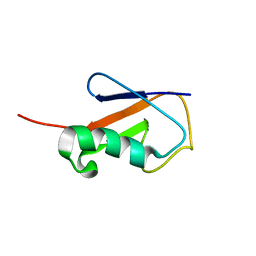 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
6KVL
 
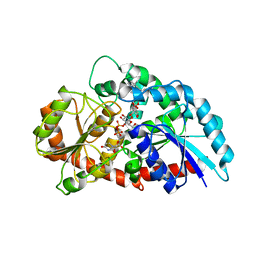 | | Crystal structure of UDP-RebB-SrUGT76G1 | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVI
 
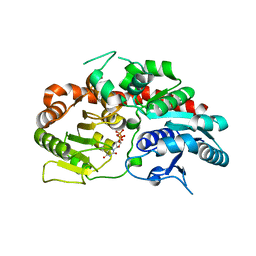 | | Crystal structure of UDP-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVJ
 
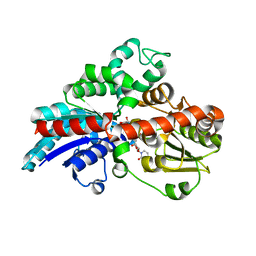 | | Crystal structure of UDPX-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVK
 
 | | Crystal structure of UDP-Sm-SrUGT76G1 | | Descriptor: | Steviolmonoside, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6HWN
 
 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
6HWM
 
 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
9BEW
 
 | |
9BER
 
 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BF6
 
 | |
1TV8
 
 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
