7XK6
 
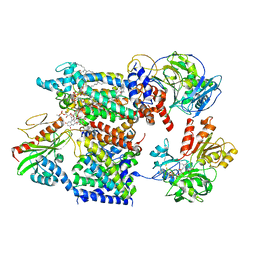 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with aurachin D-42 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aurachin D, CALCIUM ION, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK3
 
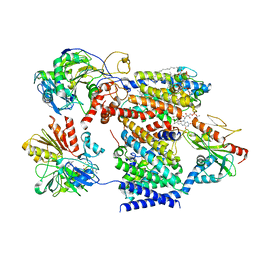 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 1 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
8JJE
 
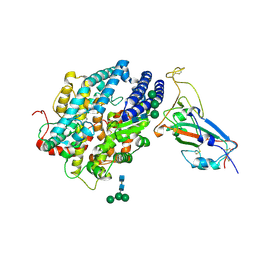 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
6DL7
 
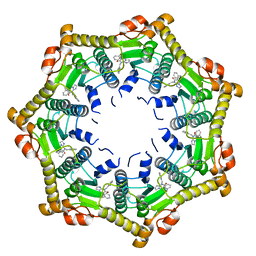 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
5H47
 
 | | Crystal structure of AOL complexed with 2-MeSe-Fuc | | Descriptor: | Uncharacterized protein, methyl 6-deoxy-2-Se-methyl-2-seleno-alpha-L-galactopyranoside | | Authors: | Kato, R, Nishikawa, Y, Makyio, H. | | Deposit date: | 2016-10-31 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of seleno-fucose compounds and their application to the X-ray structural determination of carbohydrate-lectin complexes using single/multi-wavelength anomalous dispersion phasing
Bioorg. Med. Chem., 25, 2017
|
|
7XK7
 
 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with korormicin | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
8YWT
 
 | | The isolated Vo domain of V/A-ATPase from Thermus thermophilus. | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nishida, Y, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
8YXZ
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state1 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Kishikawa, J, Nishida, Y, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
8YY0
 
 | | Vo domain of V/A-ATPase from Thermus thermophilus state2 | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit I, V-type ATP synthase, ... | | Authors: | Kishikawa, J, Nishida, Y, Nakano, A, Yokoyama, K. | | Deposit date: | 2024-04-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary mechanism of the prokaryotic V o motor driven by proton motive force.
Nat Commun, 15, 2024
|
|
6B30
 
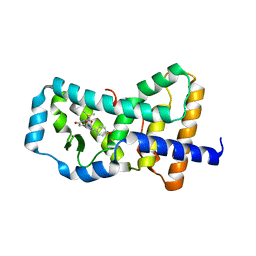 | | Structure of RORgt in complex with a novel inverse agonist 1 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-oxo-2-{[4-(trimethylsilyl)phenyl]amino}ethyl]-N-methyl-3-oxo-2,3-dihydro-1,2-oxazole-5-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of orally efficacious ROR gamma t inverse agonists, part 1: Identification of novel phenylglycinamides as lead scaffolds.
Bioorg. Med. Chem., 26, 2018
|
|
2MEE
 
 | |
5VZL
 
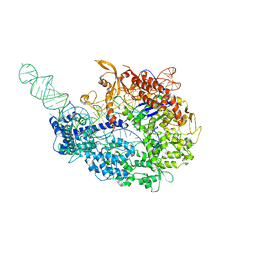 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
5YIO
 
 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
7DOJ
 
 | |
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
4UBD
 
 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LKX
 
 | | The structure of PRRSV helicase | | Descriptor: | CITRIC ACID, GLYCEROL, RNA-dependent RNA polymerase, ... | | Authors: | Shi, Y.J, Tong, X.H, Peng, G.Q. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
J.Virol., 94, 2020
|
|
5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
3SKG
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-(1,2,3,4-tetrahydro-3-isoquinolinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(3R)-1,2,3,4-tetrahydroisoquinolin-3-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-06-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Synthesis and in vivo evaluation of cyclic diaminopropane BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SHZ
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SHY
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-fluoro-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SIE
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
 | |
