4XDY
 
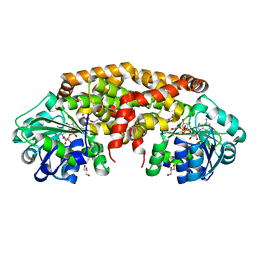 | | Structure of NADH-preferring ketol-acid reductoisomerase from an uncultured archean | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
4XEH
 
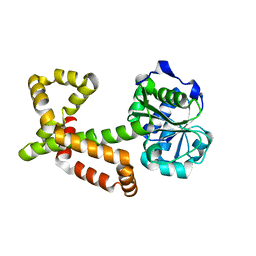 | |
7WED
 
 | |
7WEA
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEE
 
 | |
7WEB
 
 | |
7WEF
 
 | |
7WEC
 
 | |
7WE9
 
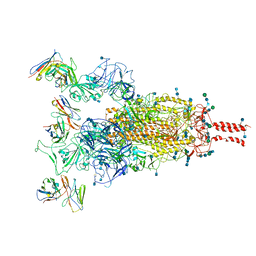 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
6US9
 
 | |
6US8
 
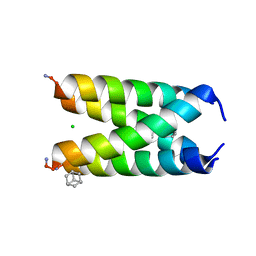 | |
7WLC
 
 | |
7WR8
 
 | |
7WRO
 
 | | Local structure of BD55-3372 and delta spike | | Descriptor: | 3372H, 3372L, Spike protein S1 | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7WRL
 
 | |
7WRZ
 
 | | Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5840H, BD55-5840L, ... | | Authors: | Zhang, Z.Z, Xiao, J.J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
3ONE
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7X6A
 
 | |
7WRJ
 
 | |
7WRY
 
 | |
1BSJ
 
 | | COBALT DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
7XNS
 
 | | SARS-CoV-2 Omicron BA.2.12.1 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
