1RZW
 
 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
3LZW
 
 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from bacillus subtilis (form I) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
6A7X
 
 | | Rat Xanthine oxidoreductase, D428A variant, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
6ADJ
 
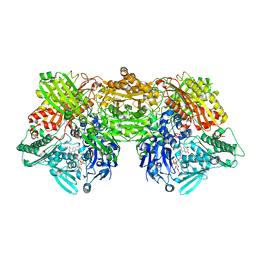 | | Rat Xanthine oxidoreductase, D428E variant | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-08-01 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
3AQX
 
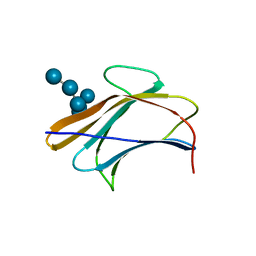 | | Crystal structure of Bombyx mori beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, GLYCEROL, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3WBD
 
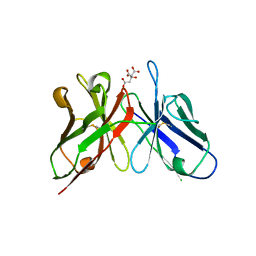 | | Crystal structure of anti-polysialic acid antibody single chain Fv fragment (mAb735) complexed with octasialic acid | | Descriptor: | CITRATE ANION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, single chain Fv fragment of mAb735 | | Authors: | Nagae, M, Ikeda, A, Hanashima, S, Kitajima, K, Sato, C, Yamaguchi, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: insight into the binding preference for polysialic acid.
J.Biol.Chem., 288, 2013
|
|
3WOG
 
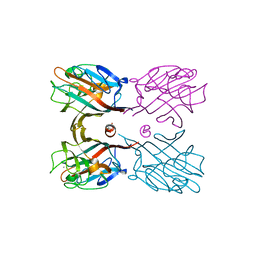 | | Crystal structure plant lectin in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-12-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3VYK
 
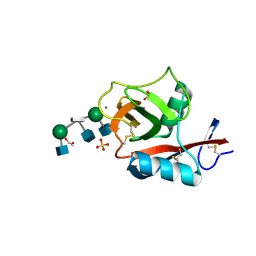 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 in complex with N-glycan | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, C-type lectin domain family 4, ... | | Authors: | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
3WCR
 
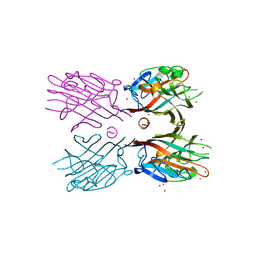 | | Crystal structure of plant lectin (ligand-free form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Erythroagglutinin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3WCS
 
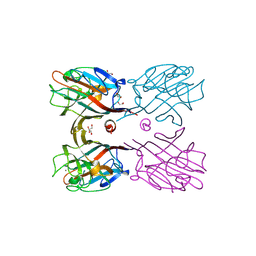 | | Crystal structure of plant lectin (ligand-bound form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
2E84
 
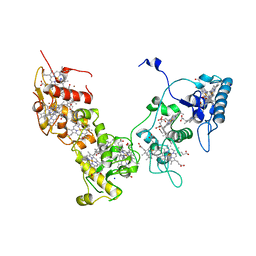 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F) in the presence of zinc ion | | Descriptor: | High-molecular-weight cytochrome c, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Shibata, N, Suto, K, Sato, M, Morimoto, Y, Kitamura, M, Higuchi, Y. | | Deposit date: | 2007-01-17 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F)
To be Published
|
|
5AVE
 
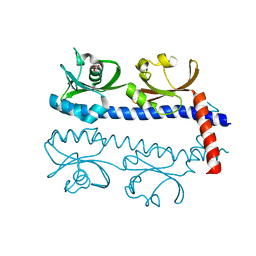 | | The ligand binding domain of Mlp37 with serine | | Descriptor: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5AWE
 
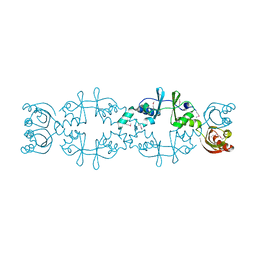 | | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains | | Descriptor: | Putative acetoin utilization protein, acetoin dehydrogenase | | Authors: | Nakabayashi, M, Shibata, N, Kanagawa, M, Nakagawa, N, Kuramitsu, S, Higuchi, Y. | | Deposit date: | 2015-07-03 | | Release date: | 2016-05-18 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Extremophiles, 20, 2016
|
|
5AV7
 
 | | Crystal structure of Calsepa lectin in complex with bisected glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, Lectin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-06-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins
Sci Rep, 6, 2016
|
|
5AVF
 
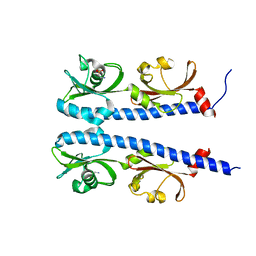 | | The ligand binding domain of Mlp37 with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
3VS1
 
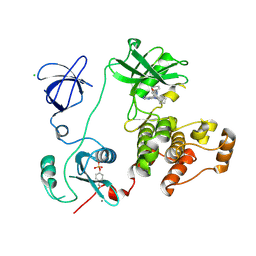 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS2
 
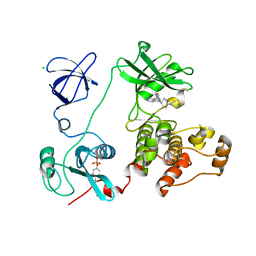 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Parker, J.L, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VRZ
 
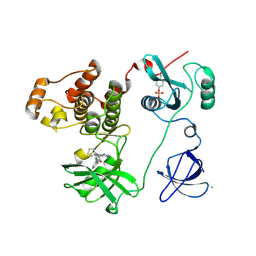 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS4
 
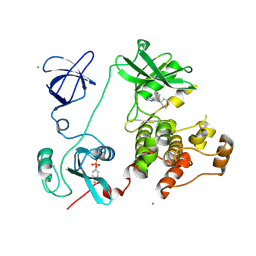 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
5AVA
 
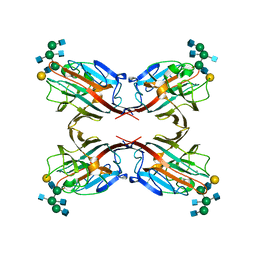 | |
3VS0
 
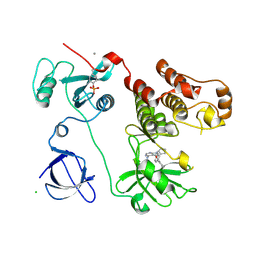 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
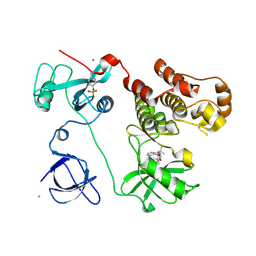 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
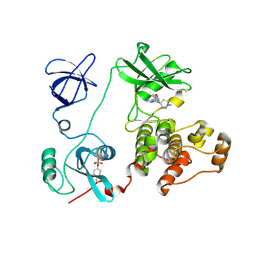 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS3
 
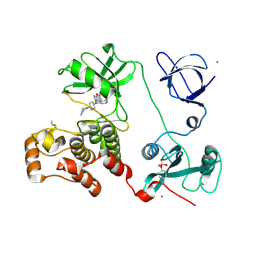 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
