2L3F
 
 | | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76 | | Descriptor: | Uncharacterized protein | | Authors: | Aramini, J.M, Hamilton, K, Ciccosanti, C.T, Wang, H, Lee, H.W, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76
To be Published
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1FUU
 
 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1LX8
 
 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
1FXV
 
 | | PENICILLIN ACYLASE MUTANT IMPAIRED IN CATALYSIS WITH PENICILLIN G IN THE ACTIVE SITE | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE, PENICILLIN G | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-27 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
1MIX
 
 | | Crystal structure of a FERM domain of Talin | | Descriptor: | Talin | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1G1P
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
2L5E
 
 | | Complex between BD1 of Brd3 and GATA-1 C-tail | | Descriptor: | Bromodomain-containing protein 3, GATA-1 | | Authors: | Gamsjaeger, R, Webb, S.R, Lamonica, J.M, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2010-10-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3.
Mol. Cell. Biol., 31, 2011
|
|
2L7M
 
 | |
1M1A
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
2L9Z
 
 | | Zinc knuckle in PRDM4 | | Descriptor: | PR domain zinc finger protein 4, ZINC ION | | Authors: | Briknarova, K, Atwater, D.Z, Glicken, J.M, Maynard, S.J, Ness, T.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PR/SET domain in PRDM4 is preceded by a zinc knuckle.
Proteins, 79, 2011
|
|
1G6N
 
 | | 2.1 ANGSTROM STRUCTURE OF CAP-CAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | Passner, J.M, Schultz, S.C, Steitz, T.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution.
J.Mol.Biol., 304, 2000
|
|
1NO6
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | Descriptor: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
2LE2
 
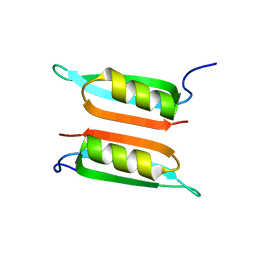 | | Novel dimeric structure of phage phi29-encoded protein p56: Insights into Uracil-DNA glycosylase inhibition | | Descriptor: | P56 | | Authors: | Asensio, J, Perez-Lago, L, Lazaro, J.M, Gonzalez, C, Serrano-Heras, G, Salas, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel dimeric structure of phage 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition.
Nucleic Acids Res., 39, 2011
|
|
2L05
 
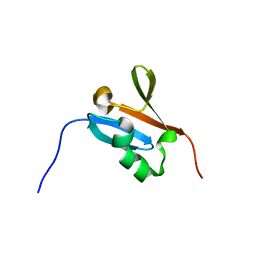 | | Solution NMR Structure of the Ras-binding domain of Serine/threonine-protein kinase B-raf from Homo sapiens, Northeast Structural Genomics Consortium Target HR4694F | | Descriptor: | Serine/threonine-protein kinase B-raf | | Authors: | Aramini, J.M, Janjua, H, Ciccosanti, C, Shastry, R, Huang, Y.J, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ras-binding domain of Serine/threonine-protein kinase B-raf from Homo sapiens, Northeast Structural Genomics Consortium Target HR4694F
To be Published
|
|
1FUK
 
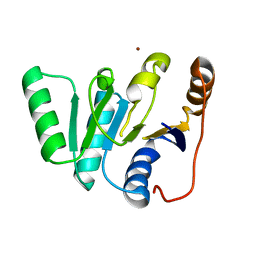 | |
1NY4
 
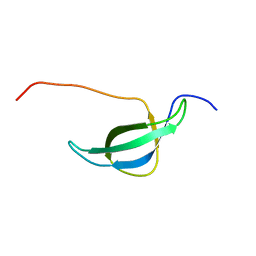 | | Solution structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Northeast Structural Genomics Consortium target JR19. | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Aramini, J.M, Cort, J.R, Huang, Y.J, Xiao, R, Acton, T.B, Ho, C.K, Shih, L.-Y, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii.
Protein Sci., 12, 2003
|
|
1NNY
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | Descriptor: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1O28
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FdUMP at 2.1 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1NT0
 
 | | Crystal structure of the CUB1-EGF-CUB2 region of MASP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Feinberg, H, Uitdehaag, J.C.M, Davies, J.M, Wallis, R, Drickamer, K, Weis, W.I. | | Deposit date: | 2003-01-28 | | Release date: | 2003-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2
Embo J., 22, 2003
|
|
2NV5
 
 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1O29
 
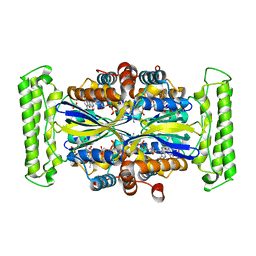 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and FdUMP at 2.0 A resolution | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1IGA
 
 | |
1NT1
 
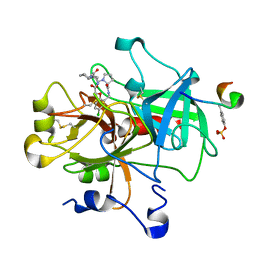 | | thrombin in complex with selective macrocyclic inhibitor | | Descriptor: | (6R,21AS)-17-CHLORO-6-CYCLOHEXYL-2,3,6,7,10,11,19,20-OCTAHYDRO-1H,5H-PYRROLO[1,2-K][1,4,8,11,14]BENZOXATETRAAZA-CYCLOHEPTADECINE-5,8,12,21(9H,13H,21AH)-TETRONE, Hirudin, thrombin | | Authors: | Nantermet, P.G, Barrow, J.C, Newton, C.L, Pellicore, J.M, Young, M, Lewis, S.D, Lucas, B.J, Krueger, J.A, McMasters, D.R, Yan, Y, Kuo, L.C, Vacca, J.P, Selnick, H.G. | | Deposit date: | 2003-01-28 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent and selective macrocyclic thrombin inhibitors
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NTJ
 
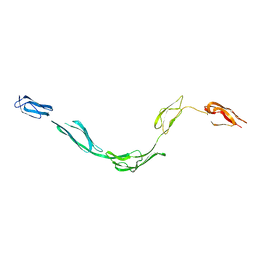 | | Model of rat Crry determined by solution scattering, curve fitting and homology modelling | | Descriptor: | complement receptor related protein | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The Extended Multidomain Solution Structures of the Complement Protein
Crry and its Chimeric Conjugate Crry-Ig by Scattering, Analytical
Ultracentrifugation and Constrained Modelling: Implications for Function and
Therapy
J.Mol.Biol., 329, 2003
|
|
