7TN1
 
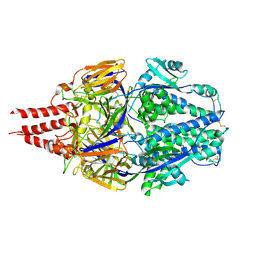 | | Multistate design to stabilize viral class I fusion proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Huang, J, Banerjee, A, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-01-20 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Nat Commun, 15, 2024
|
|
7TUR
 
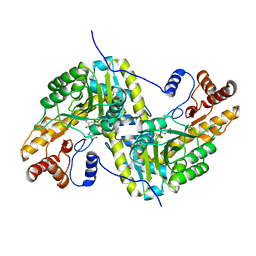 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
7TT8
 
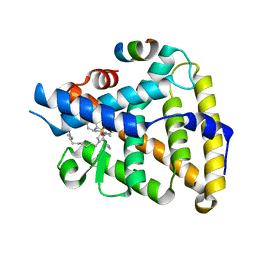 | | Human LRH-1 LBD bound to agonist 6N-10CA and fragment of Tif2 coactivator | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Modulation of Nuclear Receptor LRH-1 through Targeting Buried and Surface Regions of the Binding Pocket.
J.Med.Chem., 65, 2022
|
|
7UK2
 
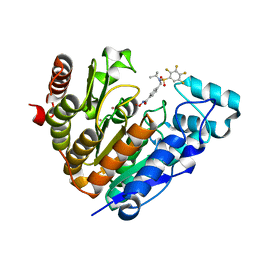 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NN-390 | | Descriptor: | Hdac6 protein, N-hydroxy-4-{[(propan-2-yl)(2,3,4,5-tetrafluorobenzene-1-sulfonyl)amino]methyl}benzamide, POTASSIUM ION, ... | | Authors: | Erdogan, F, Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Efficacy and Drug Synergy of HDAC6-Selective Inhibitor NN-429 in Natural Killer (NK)/T-Cell Lymphoma.
Pharmaceuticals, 15, 2022
|
|
7UPH
 
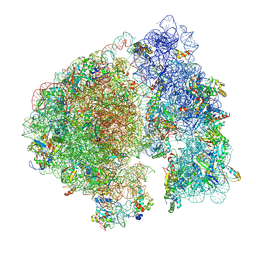 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
7Q6E
 
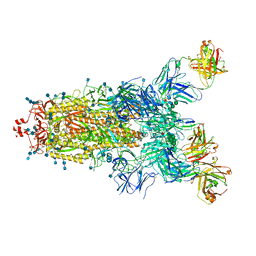 | | Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-49 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-07 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7A9I
 
 | |
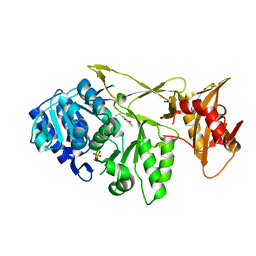
7A9J
 
 | |
7AYG
 
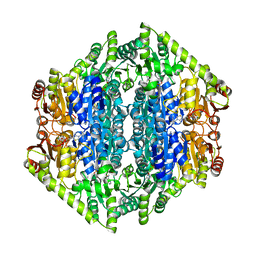 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
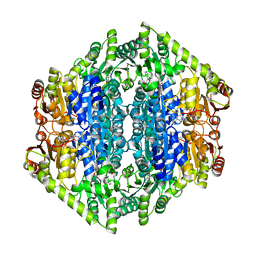 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7AM1
 
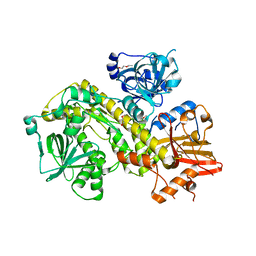 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
7BJ3
 
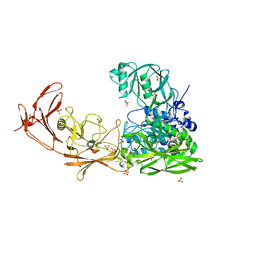 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
8CTK
 
 | |
8DSS
 
 | |
8DTA
 
 | |
8EB9
 
 | |
8EBB
 
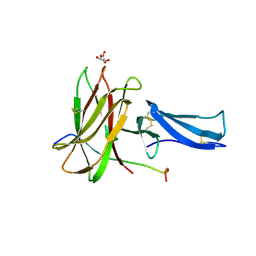 | |
8EE2
 
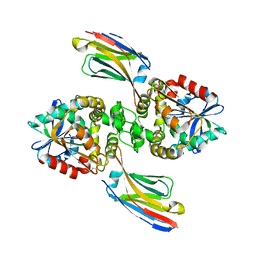 | |
8E2N
 
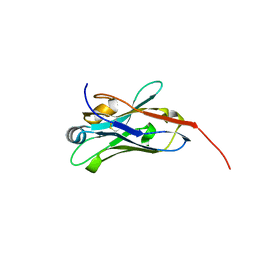 | |
8ERA
 
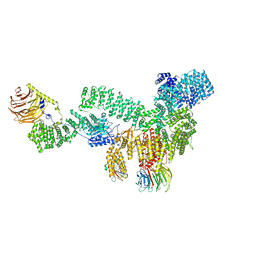 | | RMC-5552 in complex with mTORC1 and FKBP12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1-[6-{[(3M)-4-amino-3-(2-amino-1,3-benzoxazol-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-3,4-dihydroisoquinolin-2(1H)-yl]-3-hydroxypropan-1-one, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8F43
 
 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8CR8
 
 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ER6
 
 | | FKBP12-FRB in Complex with Compound 11 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
