5ZWZ
 
 | |
5ZWX
 
 | |
7DCD
 
 | |
7V4Q
 
 | |
7V4R
 
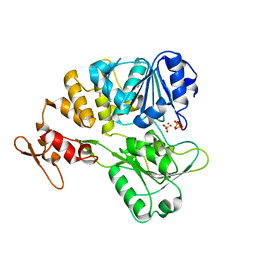 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
6BOJ
 
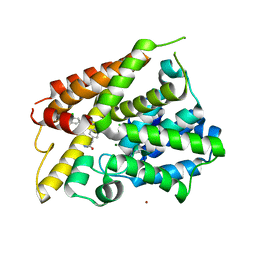 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
7Y7M
 
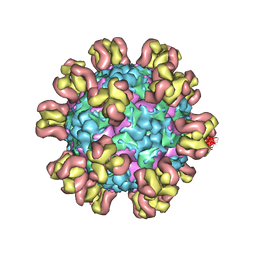 | |
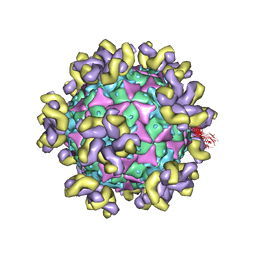
7YRH
 
 | |
7YRF
 
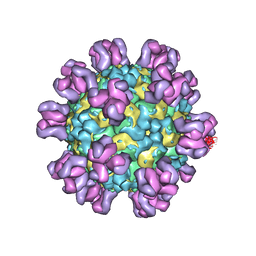 | |
7YV7
 
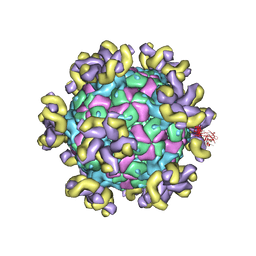 | |
7YV2
 
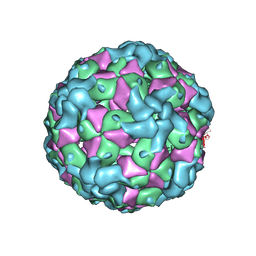 | |
7YMS
 
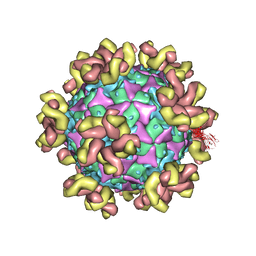 | |
7RX6
 
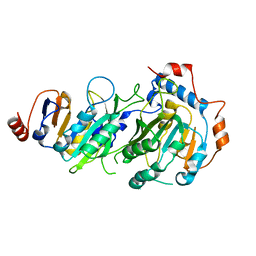 | |
7RX7
 
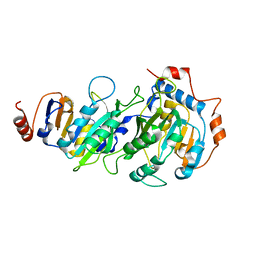 | |
7RX8
 
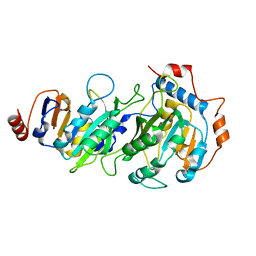 | |
5W93
 
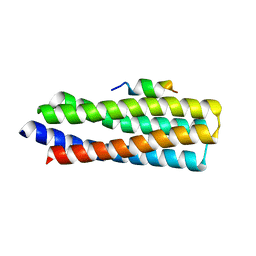 | | p130Cas complex with paxillin LD1 | | Descriptor: | Breast cancer anti-estrogen resistance protein 1, Paxillin | | Authors: | Miller, D.J, Zheng, J.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin.
J. Biol. Chem., 292, 2017
|
|
4LGH
 
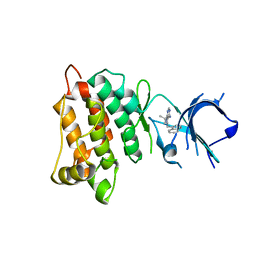 | |
4LGG
 
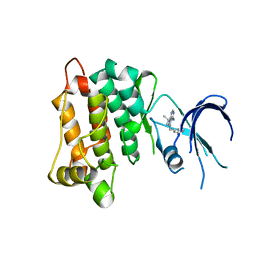 | | Structure of 3MB-PP1 bound to analog-sensitive Src kinase | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Dar, A.C, Shokat, K.M. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology.
Acs Chem.Biol., 8, 2013
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JJB
 
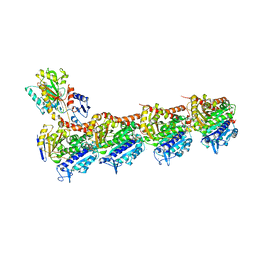 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
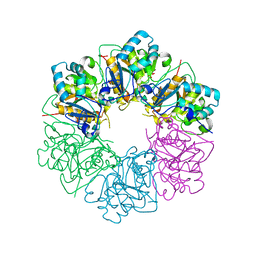
7ESV
 
 | |
7ESQ
 
 | |
7ESP
 
 | |
7ESO
 
 | |
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
