8DWG
 
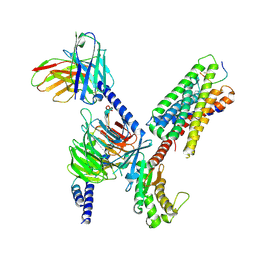 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382 | | Descriptor: | 2-[(cyclopropanesulfonyl)amino]-N-(2-ethoxyphenyl)benzamide, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
8DWC
 
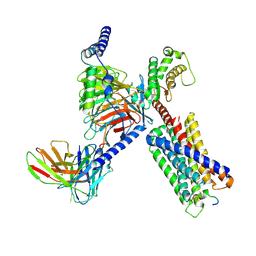 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
4XGK
 
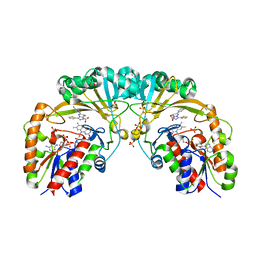 | | Crystal structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with 2-[4-(4-chlorophenyl)-7-(2-thienyl)-2-thia-5,6,8,9-tetrazabicyclo[4.3.0]nona-4,7,9-trien-3-yl]acetic | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Wangkanont, K, Heroux, A, Forest, K.T, Kiessling, L.L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents.
Acs Chem.Biol., 10, 2015
|
|
4XX9
 
 | |
3ID7
 
 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
3ITC
 
 | | Crystal structure of Sco3058 with bound citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
4JQN
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Hydroxybenzaldehyde | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, P-HYDROXYBENZALDEHYDE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMT
 
 | |
4JQJ
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinoline | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JN0
 
 | |
4JMS
 
 | |
4JPU
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with Benzamidine | | Descriptor: | BENZAMIDINE, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQM
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinazoline | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMV
 
 | |
4JMB
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 5,6,7,8-tetrahydrothieno[2,3-b]quinolin-4-amine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6,7,8-tetrahydrothieno[2,3-b]quinolin-4-amine, Cytochrome c peroxidase, ... | | Authors: | Barelier, S, Fischer, M. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Docking to a water-filled model binding site in Cytochrome c Peroxidase
To be published
|
|
4JMZ
 
 | |
4JPL
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Azaindole | | Descriptor: | 1H-pyrrolo[3,2-b]pyridine, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQK
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 2-(2-aminopyridin-1-ium-1-yl)ethanol | | Descriptor: | 2-(2-aminopyridin-1-ium-1-yl)ethanol, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JPT
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with quinazoline-2,4-diamine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
7HCB
 
 | |
7HCF
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000313 | | Descriptor: | (2R)-3-phenyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, (2S)-3-phenyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, CHLORIDE ION, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HCH
 
 | |
7HCI
 
 | |
7HCM
 
 | |
