4G3E
 
 | |
4G3F
 
 | |
3P80
 
 | |
3P82
 
 | |
3P7Y
 
 | |
3P81
 
 | |
3P74
 
 | |
2YWZ
 
 | |
2BRL
 
 | |
5C98
 
 | | 1.45A resolution structure of SRPN18 from Anopheles gambiae | | Descriptor: | AGAP007691-PB | | Authors: | Lovell, S, Battaile, K.P, Gulley, M, Zhang, X, Meekins, D.A, Gao, F.P, Michel, K. | | Deposit date: | 2015-06-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 angstrom resolution structure of SRPN18 from the malaria vector Anopheles gambiae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
2BRK
 
 | |
2Q8A
 
 | | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody | | Descriptor: | 1F9 heavy chain, 1F9 light chain, Apical membrane antigen 1 | | Authors: | Gupta, A, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2007-06-10 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody.
PLoS Pathog., 3, 2007
|
|
3DUY
 
 | | Crystal structure of human beta-secretase in complex with NVP-AFJ144 | | Descriptor: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based design and synthesis of macrocyclic peptidomimetic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2Q8B
 
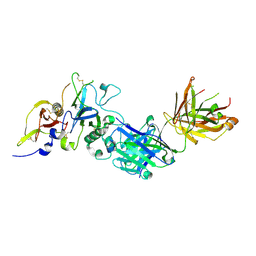 | | Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody | | Descriptor: | 1F9 heavy chain, 1F9 light chain, Apical membrane antigen 1 | | Authors: | Gupta, A, Murphy, V.J, Anders, R.F, Batchelor, A.H. | | Deposit date: | 2007-06-10 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Malaria Antigen AMA1 in Complex with a Growth-Inhibitory Antibody
Plos Pathog., 3, 2007
|
|
1N8E
 
 | | Fragment Double-D from Human Fibrin | | Descriptor: | Fibrin alpha/alpha-E chain, Fibrin beta chain, Fibrin gamma chain | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The Crystal Structure of Fragment Double-D from Cross-Linked Lamprey Fibrin Reveals Isopeptide Linkages across an Unexpected D-D Interface
Biochemistry, 41, 2002
|
|
1PSM
 
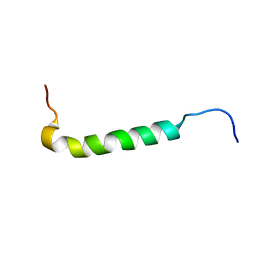 | |
1QU0
 
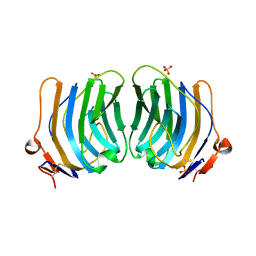 | | CRYSTAL STRUCTURE OF THE FIFTH LAMININ G-LIKE MODULE OF THE MOUSE LAMININ ALPHA2 CHAIN | | Descriptor: | CALCIUM ION, LAMININ ALPHA2 CHAIN, SULFATE ION | | Authors: | Hohenester, E, Tisi, D, Talts, J.F, Timpl, R. | | Deposit date: | 1999-07-05 | | Release date: | 1999-12-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin.
Mol.Cell, 4, 1999
|
|
1XGK
 
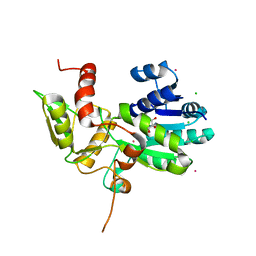 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | Descriptor: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | Authors: | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-09-17 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
3HVO
 
 | | Structure of the genotype 2B HCV polymerase bound to a NNI | | Descriptor: | 2-(3-bromophenyl)-6-[(2-hydroxyethyl)amino]-1h-benzo[de]isoquinoline-1,3(2h)-dione, Genome polyprotein | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Biological Evaluation of a Series of 1H-Benzo[de]isoquinoline-1,3(2H)-diones as Hepatitis C Virus NS5B Polymerase Inhibitors
To be Published
|
|
1LY7
 
 | | The solution structure of the the c-terminal domain of frataxin, the protein responsible for friedreich ataxia | | Descriptor: | frataxin | | Authors: | Musco, G, Stier, G, Kolmerer, B, Adinolfi, S, Martin, S, Frenkiel, T, Gibson, T, Pastore, A. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards a structural understanding of Friedreich's
ataxia: the solution structure of frataxin
Structure Fold.Des., 8, 2000
|
|
3EFR
 
 | | Biotin protein ligase R40G mutant from Aquifex aeolicus in complex with biotin | | Descriptor: | BIOTIN, Biotin [acetyl-CoA-carboxylase] ligase, SULFATE ION | | Authors: | Tron, C.M, McNae, I.W, Walkinshaw, M.D, Baxter, R.L, Campopiano, D.J. | | Deposit date: | 2008-09-10 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity.
J.Mol.Biol., 387, 2009
|
|
3EFS
 
 | | Biotin protein ligase from Aquifex aeolicus in complex with biotin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BIOTIN, Biotin [acetyl-CoA-carboxylase] ligase, ... | | Authors: | Tron, C.M, McNae, I.W, Walkinshaw, M.D, Baxter, R.L, Campopiano, D.J. | | Deposit date: | 2008-09-10 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity.
J.Mol.Biol., 387, 2009
|
|
3OCD
 
 | | Diheme SoxAX - C236M mutant | | Descriptor: | HEME C, SoxA, SoxX | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-09 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
3OA8
 
 | | Diheme SoxAX | | Descriptor: | HEME C, SULFATE ION, SoxA, ... | | Authors: | Maher, M.J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Diheme SoxAX proteins - insights into structure and function of the active site
To be Published
|
|
3FJP
 
 | | Apo structure of Biotin protein ligase from Aquifex aeolicus | | Descriptor: | Biotin [acetyl-CoA-carboxylase] ligase, SULFATE ION | | Authors: | McNae, I.W, Tron, C.M, Baxter, R.L, Walkinshaw, M.D, Campopiano, D.J. | | Deposit date: | 2008-12-15 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity.
J.Mol.Biol., 387, 2009
|
|
