6CSX
 
 | |
5CPV
 
 | |
6VWA
 
 | |
6VW9
 
 | | C-terminal regulatory domain of the chloride transporter KCC-1 from C. elegans, proteolyzed during crystallization | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, K+/Cl-Cotransporter | | Authors: | Zimanyi, C.M, Cheung, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Regulatory Cytosolic Domain of a Eukaryotic Potassium-Chloride Cotransporter.
Structure, 28, 2020
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
3QNQ
 
 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
3PJZ
 
 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
4TKS
 
 | |
3RGK
 
 | | Crystal Structure of Human Myoglobin Mutant K45R | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hubbard, S.R. | | Deposit date: | 2011-04-08 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of a recombinant human myoglobin mutant at 2.8 A resolution
J.Mol.Biol., 20, 1990
|
|
7SDM
 
 | | [U:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SD7
 
 | | [C:Hg2+:T] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDN
 
 | | [C:Ag+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.32 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDT
 
 | | [iU:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDF
 
 | | [C:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDG
 
 | | [C:Ag+:C] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*CP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDJ
 
 | | [mC:Ag+:mC] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5CM)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDQ
 
 | | [mC:Ag+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5CM)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDS
 
 | | [C:Ag+:T] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDZ
 
 | | [iU:Hg2+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SE3
 
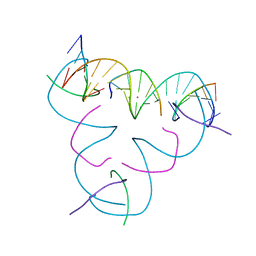 | | [iU:Hg2+:U] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SE5
 
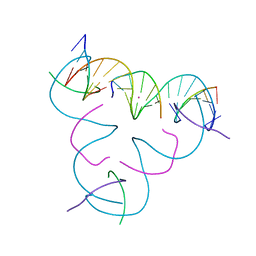 | | [iU:Hg2+:iU] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5IU)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDX
 
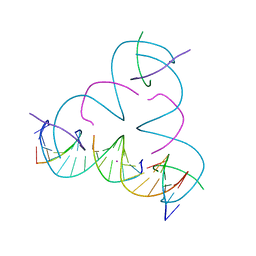 | | [U:Hg2+:mC] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SE2
 
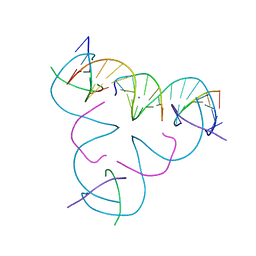 | | [C:Ag+:mC] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SDO
 
 | | [iU:Ag+:C] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5IU)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*CP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
