4AK1
 
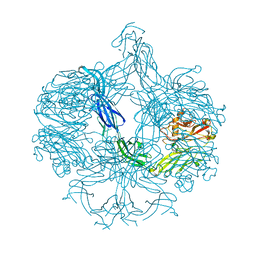 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | Descriptor: | BT_4661, SODIUM ION | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2012-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4D3L
 
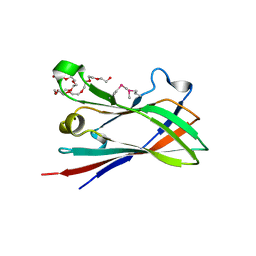 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 in the orthorhombic form | | Descriptor: | (3S)-3-HYDROXYHEPTANEDIOIC ACID, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4DH2
 
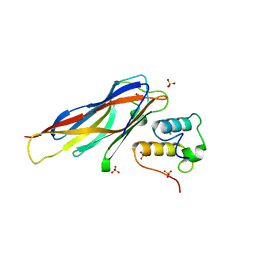 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
2VZP
 
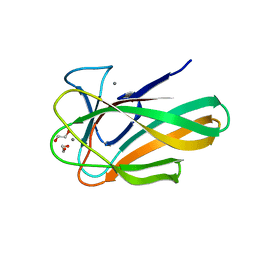 | |
2W5F
 
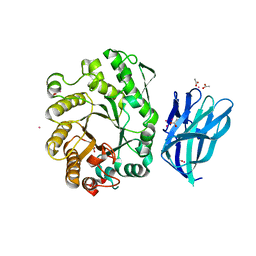 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2VZR
 
 | |
2VZQ
 
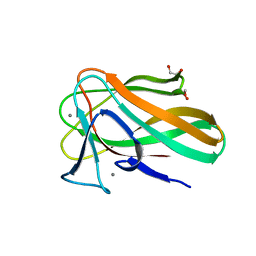 | |
2WZE
 
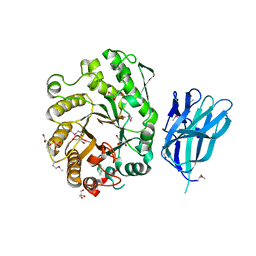 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-27 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WYS
 
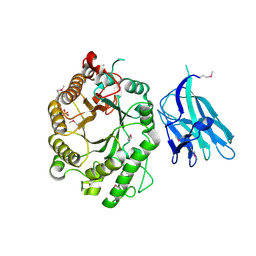 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-20 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WHK
 
 | | Structure of Bacillus subtilis mannanase man26 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Ducros, V.M.A, Davies, G.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1E5N
 
 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
