5XKD
 
 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA in complex with FMN at 2.4 angstrom | | Descriptor: | Dibenzothiophene desulfurization enzyme A, FLAVIN MONONUCLEOTIDE | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
4ES4
 
 | | Crystal structure of YdiV and FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | Authors: | Li, B, Gu, L. | | Deposit date: | 2012-04-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
3HAC
 
 | | The structure of DPP-4 in complex with piperidine fused imidazopyridine 34 | | Descriptor: | (7R,8R)-8-(2,4,5-trifluorophenyl)-6,7,8,9-tetrahydroimidazo[1,2-a:4,5-c']dipyridin-7-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminopiperidine-fused imidazoles as dipeptidyl peptidase-IV inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5XKC
 
 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA at 2.2 angstrome | | Descriptor: | Dibenzothiophene desulfurization enzyme A | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
3HAB
 
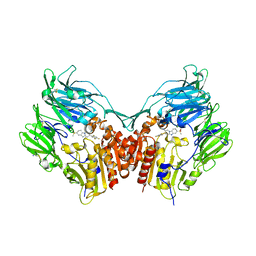 | | The structure of DPP4 in complex with piperidine fused benzimidazole 25 | | Descriptor: | (2R,3R)-7-(methylsulfonyl)-3-(2,4,5-trifluorophenyl)-1,2,3,4-tetrahydropyrido[1,2-a]benzimidazol-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopiperidine-fused imidazoles as dipeptidyl peptidase-IV inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6M05
 
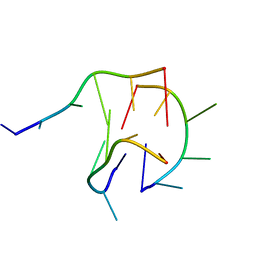 | | Trimolecular G-quadruplex | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*GP*G)-3') | | Authors: | Jing, H.T, Fu, W.Q, Zhang, N. | | Deposit date: | 2020-02-20 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance.
Nucleic Acids Res., 49, 2021
|
|
3EFK
 
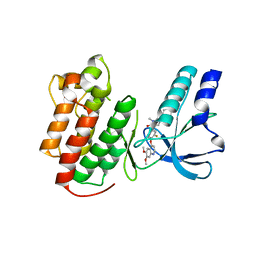 | | Structure of c-Met with pyrimidone inhibitor 50 | | Descriptor: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
7WVP
 
 | |
7WVQ
 
 | |
8KFT
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFU
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFR
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction/Ca2+ complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (25-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFW
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFS
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction complex at ground state | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
4M5F
 
 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N88
 
 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5WJK
 
 | | 2.0-Angstrom In situ Mylar structure of sperm whale myoglobin (SWMb) at 293 K | | Descriptor: | CHLORIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Broecker, J, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5WKT
 
 | | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K | | Descriptor: | Rhodopsin, SULFATE ION, Transducin Galpha peptide, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
4N80
 
 | | Crystal structure of Tse3-Tsi3 complex | | Descriptor: | CALCIUM ION, Uncharacterized protein, ZINC ION | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5Z12
 
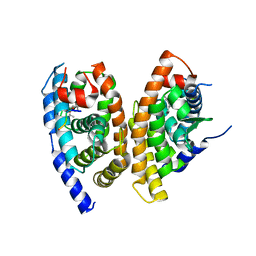 | | A structure of FXR/RXR | | Descriptor: | (9cis)-retinoic acid, 1-methylethyl 3-[(3,4-difluorophenyl)carbonyl]-1,1-dimethyl-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-12-23 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into the heterodimeric complex of the nuclear receptors FXR and RXR
J. Biol. Chem., 293, 2018
|
|
8HLO
 
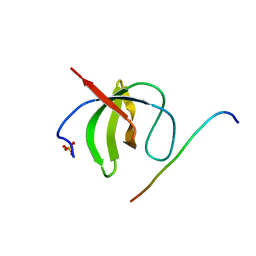 | | Crystal structure of ASAP1-SH3 and MICAL1-PRM complex | | Descriptor: | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, Proline rich motif from MICAL1, ... | | Authors: | Niu, F, Wei, Z, Yu, C. | | Deposit date: | 2022-11-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Int J Mol Sci, 24, 2023
|
|
5ECF
 
 | |
5E7X
 
 | | Ligand binding domain 1 of Penicillium marneffei MP1 protein in complex with palmitic acid | | Descriptor: | ACETATE ION, Cell wall antigen, GLYCEROL, ... | | Authors: | Lam, W.H, Zhang, H, Hao, Q. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Talaromyces marneffeiMp1 protein, a novel virulence factor, carries two arachidonic acid-binding domains to suppress inflammatory responses in hosts.
Infect. Immun., 2019
|
|
