5KNS
 
 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]hypoxanthine | | Descriptor: | (2-{[2-(6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](2-{[(E)-2-phosphonoethenyl]oxy}ethyl)amino}ethyl)phosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNP
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with [3S,4R]-(4-(Hypoxanthin-9-yl)pyrrolidin-3-yl)-oxymethanephosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
4DZU
 
 | | Complex of 3-alpha bound to gp41-5 | | Descriptor: | 3-alpha, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
4DZV
 
 | | Complex of 4-alpha/beta bound to gp41-5 | | Descriptor: | 4-alpha/beta, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
5YHH
 
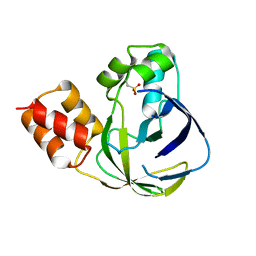 | | Crystal structure of YiiM from Geobacillus stearothermophilus | | Descriptor: | Uncharacterized conserved protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5YHI
 
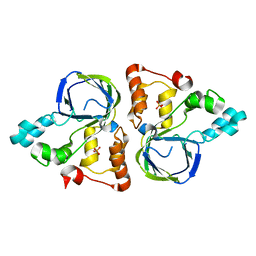 | | Crystal structure of YiiM from Escherichia coli | | Descriptor: | PHOSPHATE ION, Protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5Y8T
 
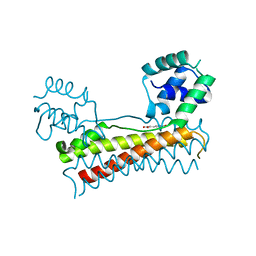 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
4DME
 
 | | GCN4 leucine zipper domain in a trimeric oligomerization state | | Descriptor: | GCN4-p1 leucine zipper domain, SULFATE ION | | Authors: | Oshaben, K.M, Salari, R, Chong, L.T, Horne, W.S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Native GCN4 Leucine-Zipper Domain Does Not Uniquely Specify a Dimeric Oligomerization State.
Biochemistry, 51, 2012
|
|
4DMD
 
 | |
4FZV
 
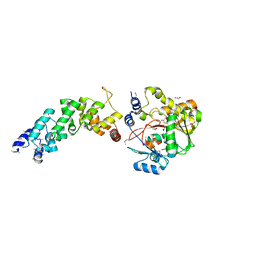 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
5ZIY
 
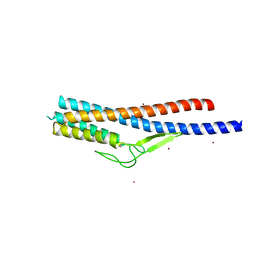 | | Crystal structure of Bacillus cereus FlgL | | Descriptor: | Flagellar hook-associated protein 3, ZINC ION | | Authors: | Hong, H.J, Kim, T.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2018-03-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FlgL and its implications for flagellar assembly
Sci Rep, 8, 2018
|
|
5Z2L
 
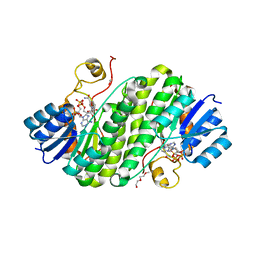 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5Y6H
 
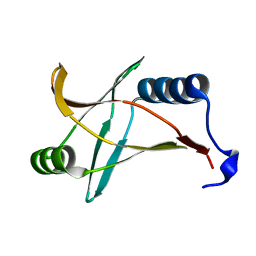 | |
5ZIZ
 
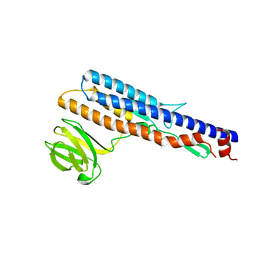 | |
5ZJ0
 
 | |
6AOZ
 
 | |
6AP0
 
 | |
6ANO
 
 | |
4H9V
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9T
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, MANGANESE (II) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-24 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9U
 
 | | Structure of Geobacillus kaustophilus lactonase, wild-type with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
1I8L
 
 | | HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC T-LYMPHOCYTE PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD80, ... | | Authors: | Stamper, C.C, Somers, W.S, Mosyak, L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses.
Nature, 410, 2001
|
|
1JCZ
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF HUMAN CARBONIC ANHYDRASE XII | | Descriptor: | ACETIC ACID, CARBONIC ANHYDRASE XII, ZINC ION | | Authors: | Whittington, D.A, Waheed, A, Ulmasov, B, Shah, G.N, Grubb, J.H, Sly, W.S, Christianson, D.W. | | Deposit date: | 2001-06-11 | | Release date: | 2001-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KV8
 
 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
1KAM
 
 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
