9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
9G6T
 
 | | p53-Y220C Core Domain Covalently Bound to 5-Chloro-6-methylpyrazine-2-carbonitrile Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranyl-6-methyl-pyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
9FHX
 
 | |
9FHV
 
 | |
9FHY
 
 | |
9FHT
 
 | |
9FI3
 
 | |
9FHU
 
 | |
9FI4
 
 | |
9FHW
 
 | |
9FHZ
 
 | |
9FI1
 
 | |
9FI5
 
 | |
9FI2
 
 | |
9FI6
 
 | |
9FI0
 
 | |
9H4A
 
 | | Ribonuclease-three Like 4 crystallization and structure determination at room temperature in the CrystalChip | | Descriptor: | Protein NUCLEAR FUSION DEFECTIVE 2 | | Authors: | Engilberge, S, Vincent, R, Coudray, L, Nilles, L, Scheer, H, Ritzenthaler, C, Sauter, C. | | Deposit date: | 2024-10-18 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallization and structure determination at room temperature in the CrystalChip.
Febs Open Bio, 15, 2025
|
|
9GBF
 
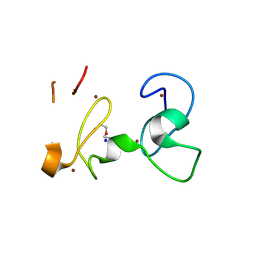 | | X-RAY structure of PHDvC5HCH tandem domain of NSD2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone-lysine N-methyltransferase NSD2, ... | | Authors: | Musco, G, Cocomazzi, P, Berardi, A, Knapp, S, Kramer, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The C-terminal PHDVC5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma.
Nucleic Acids Res., 53, 2025
|
|
9HKW
 
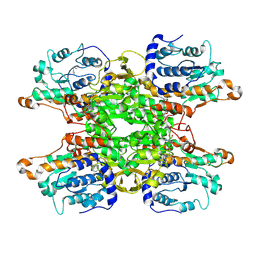 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits
To Be Published
|
|
9HKZ
 
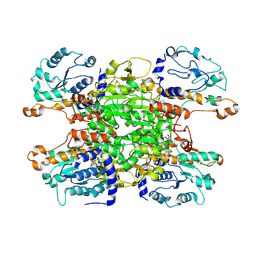 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits
To Be Published
|
|
9HKV
 
 | |
9HKY
 
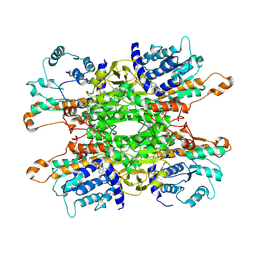 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM struture of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits
To Be Published
|
|
9HKX
 
 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits
To Be Published
|
|
9GJU
 
 | | Structure of replicating Nipah Virus RNA Polymerase Complex - RNA-bound state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphoprotein, ... | | Authors: | Sala, F, Ditter, K, Dybkov, O, Urlaub, H, Hillen, H.S. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of Nipah virus RNA synthesis.
Nat Commun, 16, 2025
|
|
9GR5
 
 | | The MKAA-RSL - sulfonato-calix[4]arene complex | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Fucose-binding lectin protein, beta-D-fructopyranose | | Authors: | Mockler, N.M, Ramberg, K.O, Flood, R.J, Crowley, P.B. | | Deposit date: | 2024-09-10 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | N-Terminal Protein Binding and Disorder-to-Order Transition by a Synthetic Receptor.
Biochemistry, 64, 2025
|
|
