3BS4
 
 | | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide | | Descriptor: | Uncharacterized protein PH0321, Unknown peptide | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide.
To be Published
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3C9F
 
 | | Crystal structure of 5'-nucleotidase from Candida albicans SC5314 | | Descriptor: | 5'-nucleotidase, FORMIC ACID, SODIUM ION, ... | | Authors: | Patskovsky, Y, Romero, R, Gilmore, M, Eberle, M, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 5'-nucleotidase from Candida albicans.
To be Published
|
|
3CBW
 
 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
2XTB
 
 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
1SSF
 
 | | Solution structure of the mouse 53BP1 fragment (residues 1463-1617) | | Descriptor: | Transformation related protein 53 binding protein 1 | | Authors: | Charier, G, Couprie, J, Alpha-Bazin, B, Meyer, V, Quemeneur, E, Guerois, R, Callebaut, I, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2004-03-24 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Tudor Tandem of 53BP1; A New Structural Motif Involved in DNA and RG-Rich Peptide Binding
Structure, 12, 2004
|
|
5MMK
 
 | | HYL-20 | | Descriptor: | GLY-ILE-LEU-SER-SER-LEU-TRP-LYS-LYS-LEU-LYS-LYS-ILE-ILE-ALA-LYS | | Authors: | Hexnerova, R. | | Deposit date: | 2016-12-10 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides.
Pathog Dis, 75, 2017
|
|
5MML
 
 | | HYL-20k | | Descriptor: | GLY-ILE-LEU-SER-SER-LEU-TRP-LYS-LYS-LEU-LYS-LYS-ILE-ILE-ALA-LYS | | Authors: | Hexnerova, R. | | Deposit date: | 2016-12-10 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides.
Pathog Dis, 75, 2017
|
|
6ZP7
 
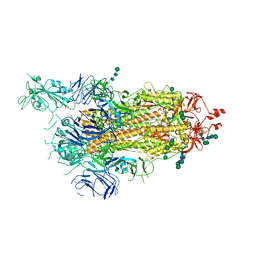 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZOW
 
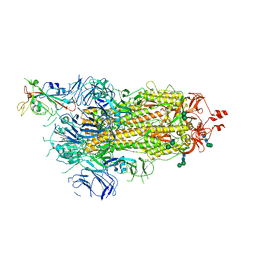 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZP5
 
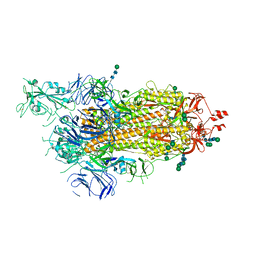 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
2M6Q
 
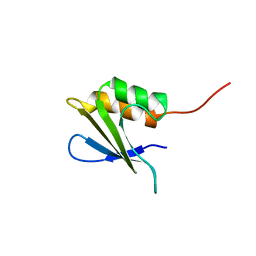 | | Refined Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Strucutral Genomics Consortium Target ZR18 | | Descriptor: | SAV1430 | | Authors: | Baran, M.C, Aramini, J.M, Huang, Y.J, Xiao, R, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M8W
 
 | | Restrained CS-Rosetta Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Structural Genomics Target ZR18. Structure determination | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M8X
 
 | | Restrained CS-Rosetta Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
3IIY
 
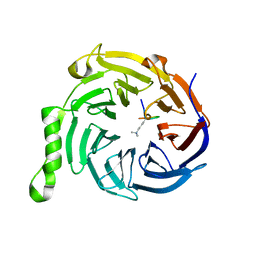 | | Crystal structure of Eed in complex with a trimethylated histone H1K26 peptide | | Descriptor: | Histone H1K26 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IIW
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K27 peptide | | Descriptor: | Histone H3 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ0
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K9 peptide | | Descriptor: | Histone H3K9 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJC
 
 | | Crystal structure of Eed in complex with NDSB-195 | | Descriptor: | ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ1
 
 | | Crystal structure of Eed in complex with a trimethylated histone H4K20 peptide | | Descriptor: | Histone H4K20 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
1PUL
 
 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | Descriptor: | Hypothetical protein C32E8.3 in chromosome I | | Authors: | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
2H73
 
 | |
