7FQN
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000497a | | Descriptor: | (1S,4R,5S,6R)-2-(methylsulfonyl)-2-azabicyclo[3.3.1]nonane-4,6-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRS
 
 | | PanDDA analysis group deposition of ground-state model of PTP1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRJ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z2856434770 | | Descriptor: | 1-(3-chlorophenyl)-N-methylmethanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
3SBN
 
 | | trichovirin I-4A in polar environment at 0.9 Angstroem | | Descriptor: | ACETONITRILE, METHANOL, Trichovirin I-4A | | Authors: | Gessmann, R, Axford, D, Petratos, K. | | Deposit date: | 2011-06-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Four complete turns of a curved 310-helix at atomic resolution: The crystal structure of the peptaibol trichovirin I-4A in polar environment suggests a transition to alpha-helix for membrane function
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JGZ
 
 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JGY
 
 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
1R03
 
 | | crystal structure of a human mitochondrial ferritin | | Descriptor: | MAGNESIUM ION, mitochondrial ferritin | | Authors: | Corsi, B, Santambrogio, P, Arosio, P, Levi, S, Langlois d'Estaintot, B, Granier, T, Gallois, B, Chevallier, J.M, Precigoux, G. | | Deposit date: | 2003-09-19 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure and Biochemical Properties of the Human Mitochondrial Ferritin and its Mutant Ser144Ala
J.Mol.Biol., 340, 2004
|
|
8OEI
 
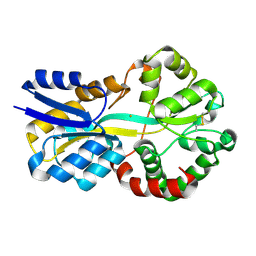 | | SFX structure of FutA after an accumulated dose of 350 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OEM
 
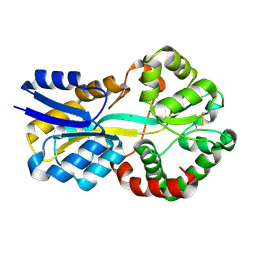 | | Crystal structure of FutA bound to Fe(II) | | Descriptor: | FE (II) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OGG
 
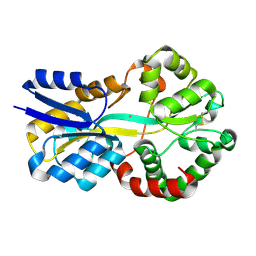 | | Crystal structure of FutA after an accumulated dose of 5 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3KTT
 
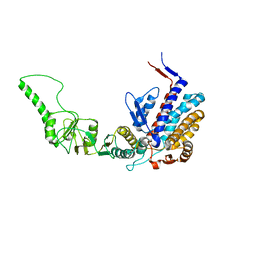 | | Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit beta | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7QZH
 
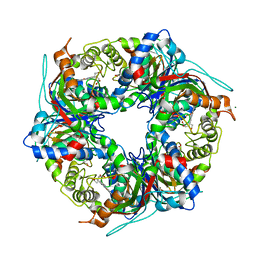 | | SFX structure of dye-type peroxidase DtpB D152A variant in the ferric state | | Descriptor: | Dyp-type peroxidase family, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
7QZE
 
 | | SFX structure of dye-type peroxidase DtpB D152A variant in the ferryl state | | Descriptor: | Dyp-type peroxidase family, MAGNESIUM ION, OXYGEN ATOM, ... | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
7QZG
 
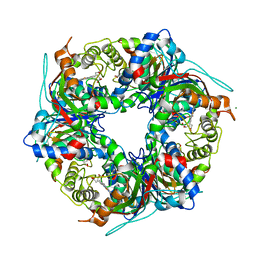 | | SFX structure of dye-type peroxidase DtpB N245A variant in the ferric state | | Descriptor: | Dyp-type peroxidase family, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
3TYD
 
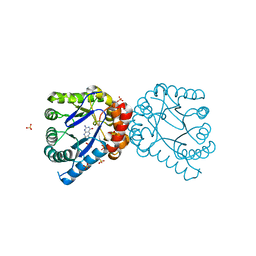 | | Dihydropteroate Synthase in complex with PPi and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, PYROPHOSPHATE 2-, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
8BCK
 
 | |
8BC5
 
 | |
8BBT
 
 | |
8BCL
 
 | |
8C4Y
 
 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RK1
 
 | | Crystal structure of FutA bound to Fe(III) solved by neutron diffraction | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-12-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (2.095 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6YRJ
 
 | | SFX structure of dye-type peroxidase DtpB in the ferric state | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative iron-dependent peroxidase | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6YRD
 
 | | SFX structure of dye-type peroxidase DtpB in the ferryl state | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Axford, D.A, Owen, R.L, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1OEV
 
 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
