5VOV
 
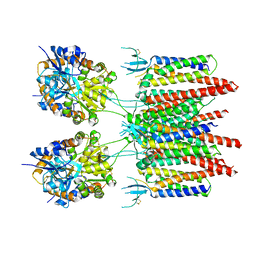 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5IZA
 
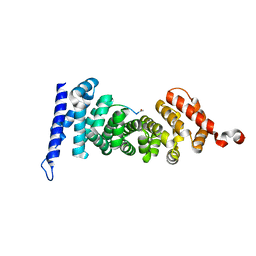 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
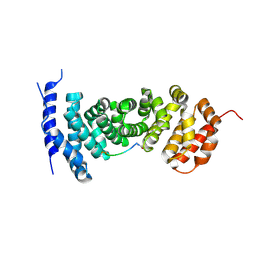 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
1PTY
 
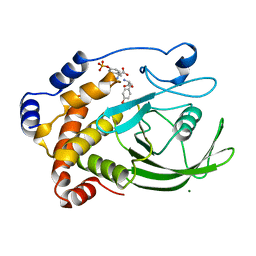 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO PHOSPHOTYROSINE MOLECULES | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Zhao, Y, Puius, Y.A, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6YSK
 
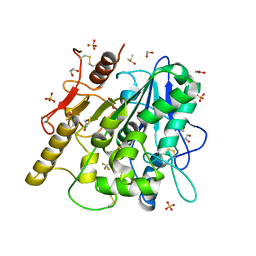 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
9BW8
 
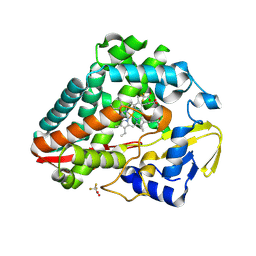 | | Structure of P450Blt from Micromonospora sp. MW-13 in Complex with Fluorinated Biarylitide | | Descriptor: | Cytochrome P450-SU1, Fluorinated Biarylitide, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hansen, M.H, Cryle, M.J, Zhao, Y. | | Deposit date: | 2024-05-21 | | Release date: | 2024-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Loss of fluorine during crosslinking by the biarylitide P450 Blt proceeds due to restricted peptide orientation.
Chem.Commun.(Camb.), 60, 2024
|
|
7TRG
 
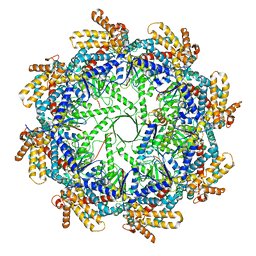 | | The beta-tubulin folding intermediate I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTN
 
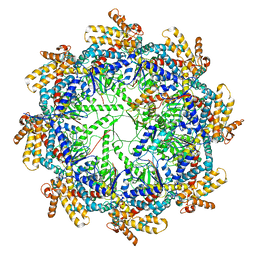 | | The beta-tubulin folding intermediate II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TUB
 
 | | The beta-tubulin folding intermediate IV | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-02 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTT
 
 | | The beta-tubulin folding intermediate III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9J7N
 
 | | Cryo-EM structure of TauT | | Descriptor: | BETA-ALANINE, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7M
 
 | | Cryo-EM structure of TauT | | Descriptor: | 2-AMINOETHANESULFONIC ACID, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7O
 
 | | Cryo-EM structure of TauT | | Descriptor: | CHLORIDE ION, CHOLESTEROL, HEXADECANE, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
6LPM
 
 | |
8SLN
 
 | |
8SLM
 
 | | Crystal structure of Deinococcus geothermalis PprI | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Zn dependent hydrolase fused to HTH domain, ... | | Authors: | Zhao, Y, Lu, H. | | Deposit date: | 2023-04-23 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA.
Nat Commun, 15, 2024
|
|
5F56
 
 | | Structure of RecJ complexed with DNA and SSB-ct | | Descriptor: | ALA-ASP-LEU-PRO-PHE, DNA (5'-D(*CP*TP*GP*AP*TP*GP*GP*CP*A)-3'), MANGANESE (II) ION, ... | | Authors: | Zhao, Y, Hua, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7MPI
 
 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N8B
 
 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
8JCN
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 58 | | Descriptor: | 1-[3-(diphenoxyphosphorylamino)phenyl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
