5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
4F8T
 
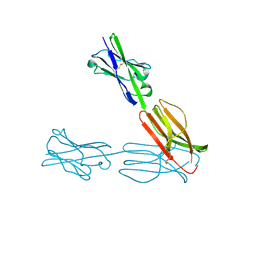 | | Crystal Structure of the Human BTN3A3 Ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A3 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell response by CD277/Butryophilin (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F9L
 
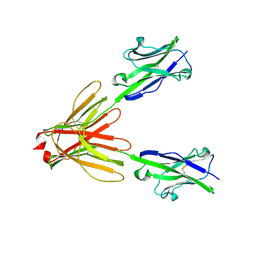 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 20.1 Single Chain Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 20.1 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1404 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F8Q
 
 | | Crystal Structure of the Human BTN3A2 Ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A2 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3789 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F9P
 
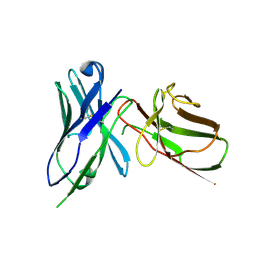 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 103.2 Single Chain Antibody | | Descriptor: | 103.2 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.519 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F80
 
 | | Crystal Structure of the human BTN3A1 ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-16 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
5YIO
 
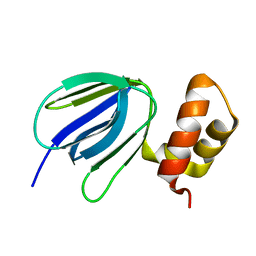 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
4XH3
 
 | |
5OLT
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Vella, P, Polyakova, A, Lindqvist, Y, Schnell, R, Bourenkov, G, Schneider, T, Schneider, G. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
5OJH
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRATE ANION, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Schneider, G, Vella, P, Lindqvist, Y, Schnell, R. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
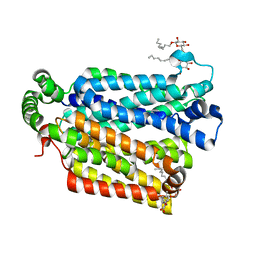 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
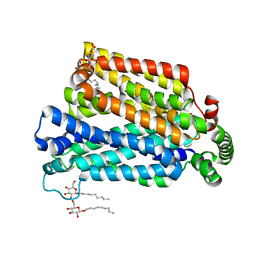 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
5GYL
 
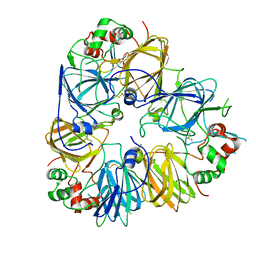 | | Structure of Cicer arietinum 11S gloubulin | | Descriptor: | legumin-like protein | | Authors: | Zhou, A, Zhang, F. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and crystallographic studies of a novel chickpea 11S globulin.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7FAP
 
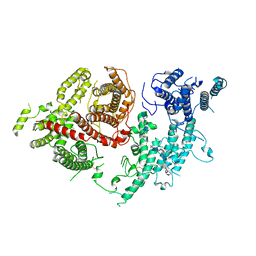 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
5DUP
 
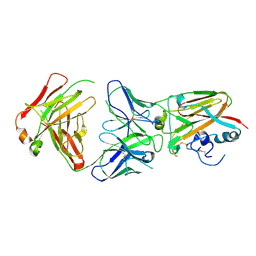 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody AVFluIgG03 | | Descriptor: | AVFluIgG03 Heavy Chain, AVFluIgG03 Light Chain, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUT
 
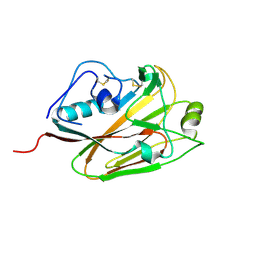 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUM
 
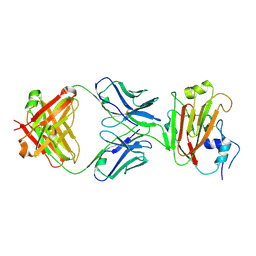 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | Authors: | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | Deposit date: | 2015-09-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUR
 
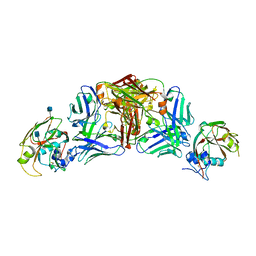 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
7RPV
 
 | |
4LKO
 
 | | Crystal structure of human DPP-IV in complex with BMS-744891 | | Descriptor: | 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-6,7-dihydro-5H-pyrrolo[3,4-b]pyridin-5-one, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-07-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
7S4S
 
 | |
5Y4L
 
 | | PRRSV nsp4 | | Descriptor: | Non-structural protein | | Authors: | Shi, Y.J, Gang, Y, Peng, G.Q. | | Deposit date: | 2017-08-04 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus.
Vet. Microbiol., 213, 2018
|
|
3E88
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8C
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor peptide | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
